FABP4-mediated lipid accumulation and lipolysis in tumor-associated macrophages promote breast cancer metastasis
Figures
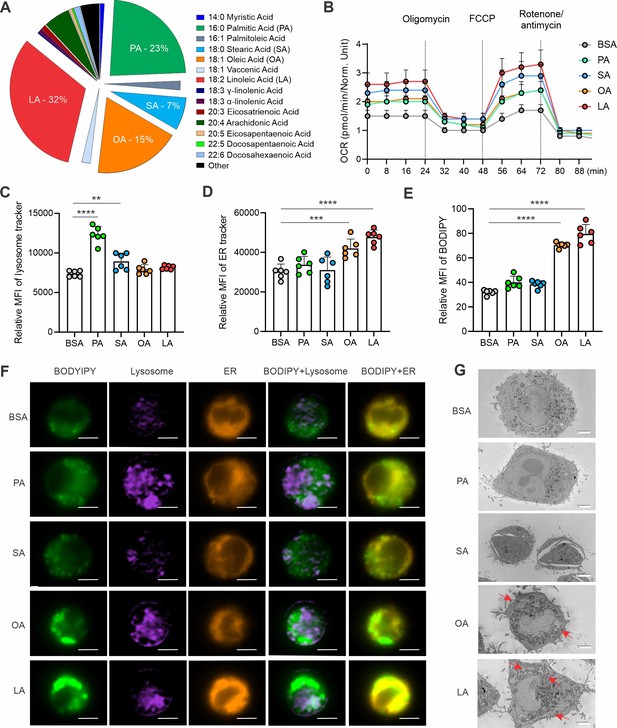
Unsaturated FAs form lipid accumulation in macrophages.
(A) Pie chart showing main compositions of free fatty acids in the serum of healthy humans. (B) Measurement of oxygen consumption rate (OCR) in macrophages treated with 200 μM of PA, SA, OA, LA or control BSA, respectively, under basal conditions or following the addition of oligomycin, FCCP or the electron transport inhibitor Rotenone/antimycin by a seahorse XF-96 analyzer (n=5). (C–E) Macrophages were treated with 200 μM of PA, SA, OA, LA or BSA for 4 hr. Flow cytometric analysis of lysosome, ER and lipid droplet formation by measuring mean fluorescent intensity (MFI) of lysosome tracker (C), ER tracker (D) and BODIPY (E) in macrophages. (F) Multispectral imaging analysis of BODIPY (green), Lysosome (purple), ER (orange), and merged images showing the colocalization status of BODIPY/lyso-tracker and BODIPY/ER-tracker in macrophages treated with PA, SA, OA, LA, or BSA at 200 μM for 4 hr. Scale bar: 7μM. (G) Analysis of lipid droplet formation (red arrow) in macrophages treated with BSA, PA, SA, OA, and LA by transmission electron microscope. Data are shown as mean ± SD in panel B-E (** p≤0.01, *** p≤0.001, **** p≤0.0001, as compared to the control BSA group, unpaired Student t test).
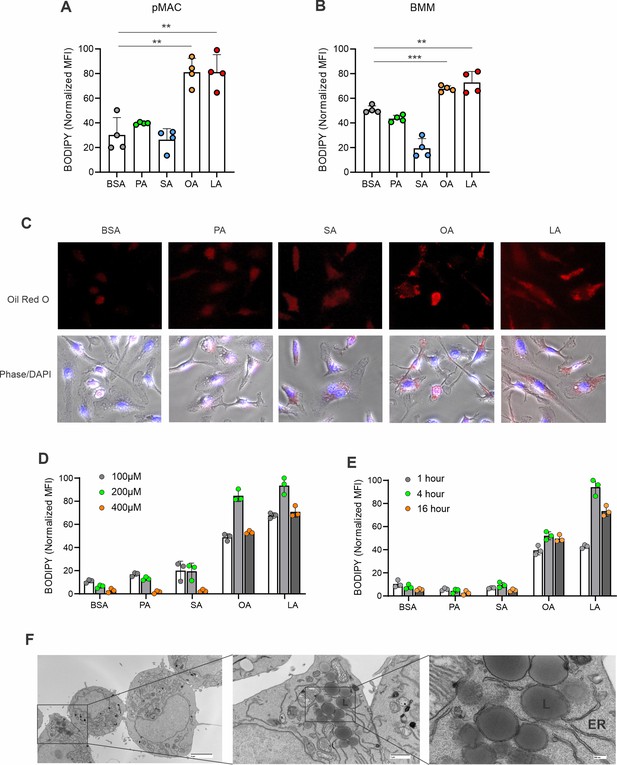
Unsaturated FA form lipid droplets in different macrophages.
(A) Flow cytometric analysis of lipid droplet formation in peritoneal macrophages treated with BSA and indicated dietary FA (100 μM). (B) Flow cytometric analysis of lipid droplet formation in bone-marrow-derived macrophages (BMM) treated with BSA and indicated dietary FA (100 μM). (C) Confocal microscopy analysis of Oil Red O staining in BMM treated with 200 μM of PA, SA, OA, LA, and control BSA for 4 hr. (D) Titration of different indicated concentrations of dietary FA in forming lipid droplets in macrophage cell lines. (E) Measurement of lipid droplet formation in macrophages treated with indicated time periods of different dietary FAs (100 μM). (F) Transmission electron microscope (TEM) images showing that LA-induced lipid droplets were formed in endoplasmic reticulum (ER) in macrophages. Data are shown as mean ± SD in panel (** p≤0.01, *** p≤0.001, unpaired Student t test).
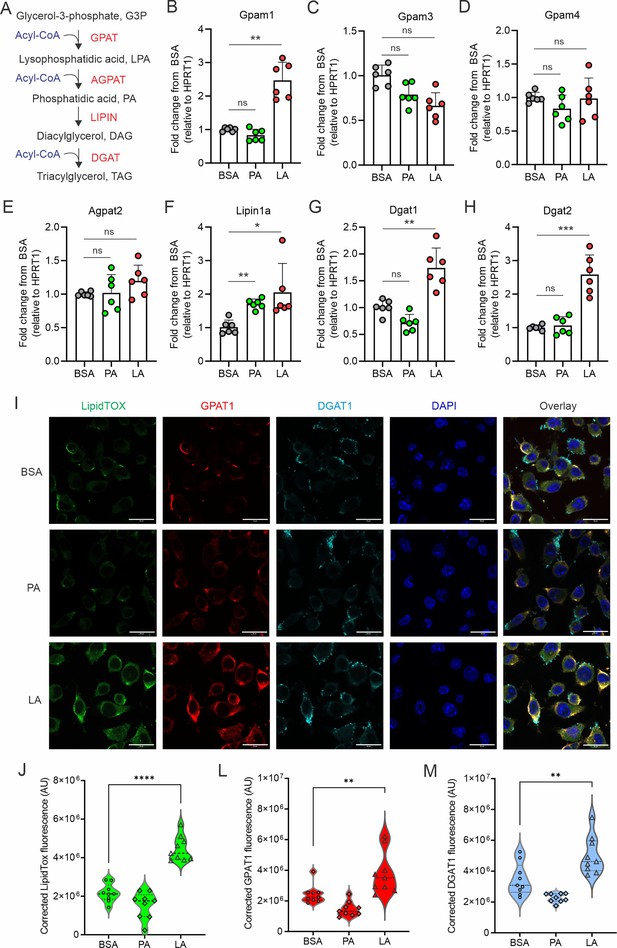
LA induces the expression of key enzymes of triacylglycerol synthesis in macrophages.
(A) Key enzymes in the biosynthesis of triacylglycerol. (B–H) Analyzing the expression of genes encoding key enzymes, including Gpam1 (B), Gpam3 (C), Gpam4 (D), Agpat2 (E), Lipin1a (F), Dgat1 (G), Dgat2 (H), in the triacylglycerol biosynthesis pathway in macrophages treated with 400 μM of PA, LA or BSA for 4 hr by real-time PCR. (I) Representative confocal images of lipid accumulation by LipidTOX staining (green), expression of GPAT1 (red), DGAT1 in macrophages treated with BSA, PA, and LA (400 μM) overnight. (J–M) Quantification of lipid accumulation (J), protein levels of GAPT1 (K) and DGAT1 (M) in macrophages treated with BSA, PA, or LA (400 μM) overnight. Data are shown as mean ± SD in panel B-H, J-M (** p≤0.01, *** p≤0.001, **** p≤0.0001, ns, non-significant, as compared to the control BSA group, unpaired Student t test).
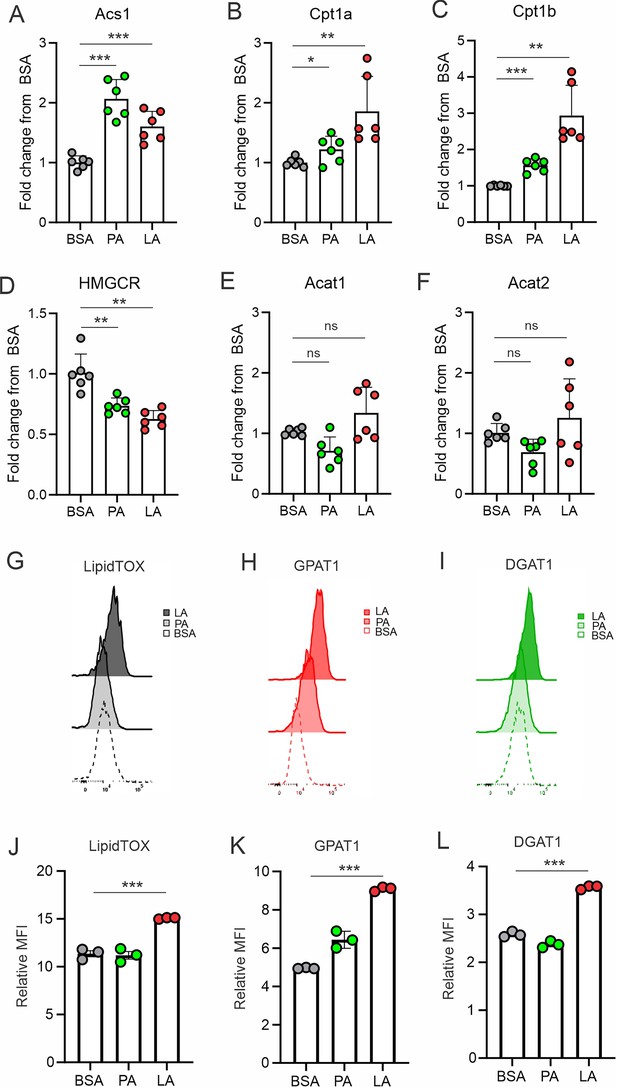
LA induces key enzyme expression in triglyceride biosynthesis.
(A–F) Analysis of FA metabolism-related genes, including Acs1 (A), Cpt1a (B), Cpt1b (C), HMGCR (D), Acat1 (E), Acat2 (F) in macrophages treated with BSA, PA or LA (400 μM) for 4 hr. (G–I) Flow cytometric staining for LipidTOX (G), GPAT1 (H), and DGAT1 (I) expression in macrophages treated with BSA, PA, or LA (400 μM). (J–L) Quantification of LipiTOX (J), GPAT1 (K), and DGAT1 (L) expression by mean fluorescent intensity (MFI) in macrophages treated with BSA, PA, or LA (400 μM). Data are shown as mean ± SD in panel A-F and J-L (* p≤0.05, ** p≤0.01, **** p≤0.0001, ns, non-significant, unpaired Student t test).
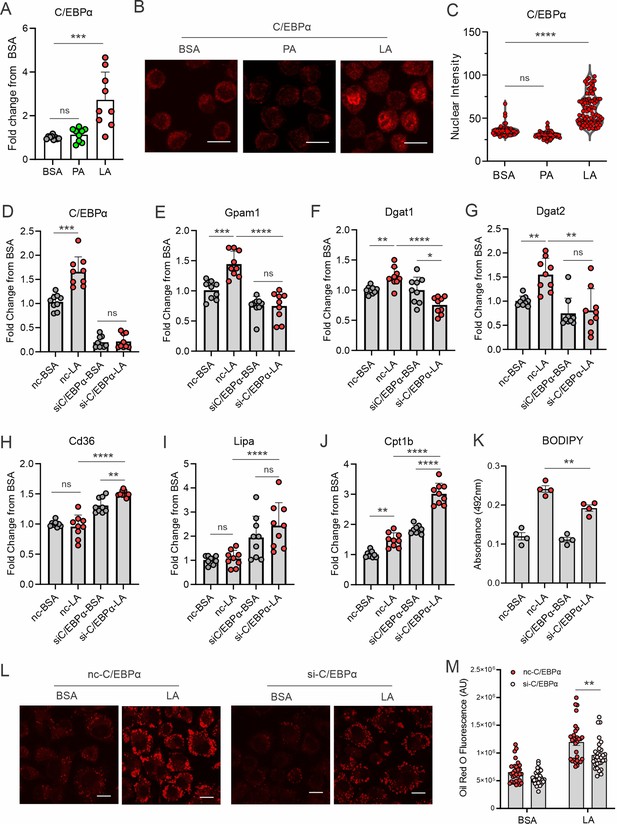
LA induces lipid accumulation through activating the C/EBPα pathway.
(A) Measurement of C/EBPα gene expression levels in macrophages treated with BSA, PA, or LA (400 μM) for 4 hr by real-time PCR. (B) Representative confocal images of C/EBPα protein expression (red) in macrophages treated with BSA, PA, or LA overnight. (C) Quantification of C/EBPα nuclear expression in macrophages treated with BSA, PA, or LA overnight by Image J analysis. (D–J) Real-time PCR analysis of the levels of C/EBPα (D), Gpam1 (E), Dgat1 (F), Dgat2 (G), Cd36 (H), Lipa (I), and Cpt1b (J) in macrophages transfected with 40 nM C/EBPα siRNA or control siNC and then treated with BSA or LA for 4 hr. (K) Measurement of BODIPY fluorescent intensity in C/EBPα-silencing or control macrophages treated with BSA or LA using a fluorescence spectroscopy. (L) Representative confocal images of Oil Red O staining in C/EBPα-silencing or control macrophages treated with BSA or LA (bar, 10 μM). (M) Quantification of Oil Red O fluorescence intensity in C/EBPα-silencing or control macrophages treated with BSA or LA. Data are shown as mean ± SD in panel A, C-K and M (*p≤0.05, ** p≤0.01, *** p≤0.001, **** p≤0.0001, ns, non-significant, as compared to the control BSA group or control siNC group, unpaired Student t test).
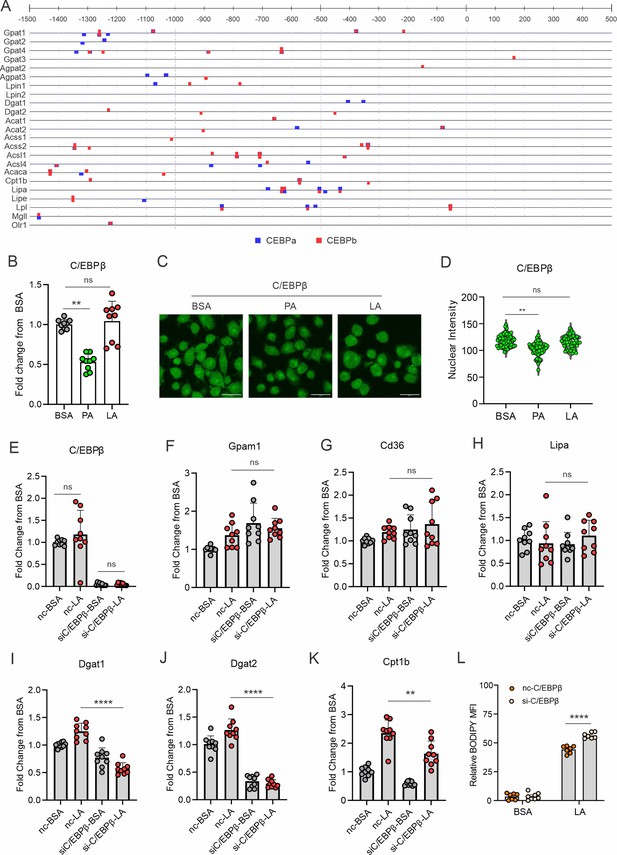
LA-induced lipid accumulation in macrophages was undependable on CEBPβ.
(A) CiiiDER software package was used to identify potential C/EBPα and C/EBPβ transcription factor binding sites in regulatory regions of relevant lipid droplet and lipolysis genes. (B) Real-time PCR analysis of CEBPβ expression macrophages treated with BSA, PA, and LA for 4 hr. (C, D) Confocal analysis for CEBPβ staining (green) in macrophages treated with BSA, PA, and LA. Quantification of CEBPβ staining is shown in panel D. (E–K) Macrophages was transfected with CEBPβ or control siRNA, and then treated with BSA or LA for 4 hr. Expression of CEBPβ (E), lipid droplet formation and other FA-metabolism related genes, including Gpam1 (F), Cd36 (G), Lipa (H), Dgat1 (I), Dgat2 (J), Cpt1b (K) was determined by real-time PCR analysis. (L) Measurement of lipid droplet formation by BODIPY staining in CEBPβ-silencing macrophages treated with BSA or LA. Data are shown as mean ± SD in panel L-O (** p≤0.01, **** p≤0.0001, ns, non-significant, unpaired Student t test).
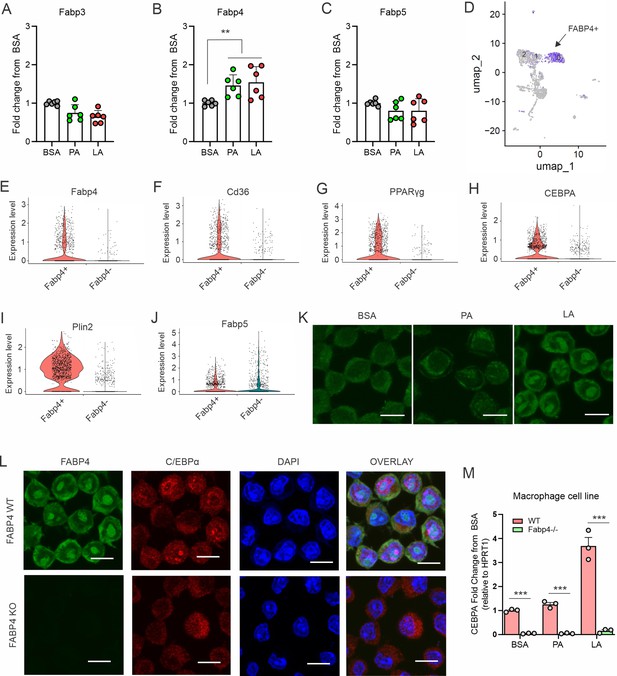
FABP4 mediates LA-induced C/EBPα expression in macrophages.
(A–C) Analysis of the expression of FABP family members, including Fabp3 (A), Fabp4 (B) and Fabp5 (C), in macrophages treated with BSA, PA or LA (400 μM) for 4 hours. (D) UMAP of FABP4-positive macrophage subsets using mouse spleen single-cell RNA sequence analysis. (E–J) Violin plots showing relative expression levels of genes, including Fabp4(E), Cd36 (F), PPARγ (G), CEBPA (H), Plin2 (I) and Fabp5 (J) between Fabp4 +vs Fabp4- macrophages indicated in (C). (K) Confocal analysis of FABP4 expression in macrophages treated with BSA, PA or LA (400 μM) (bar, 10 μM). (L) Comparison of the expression of FABP4 and C/EBPα between FABP4 wildtype (WT) and knockout (KO) macrophages in response to LA treatment (400 μM) (bar, 10 μM). (M) Realtime PCR analysis of CEBPA expression in WT and FABP4-/- macrophages treated with BSA, PA, and LA (400 μM). Data are shown as mean ± SD in panel A and L (** p≤0.01, *** p≤0.001, as compared to the control BSA group or FABP4-/- group, unpaired Student t test).
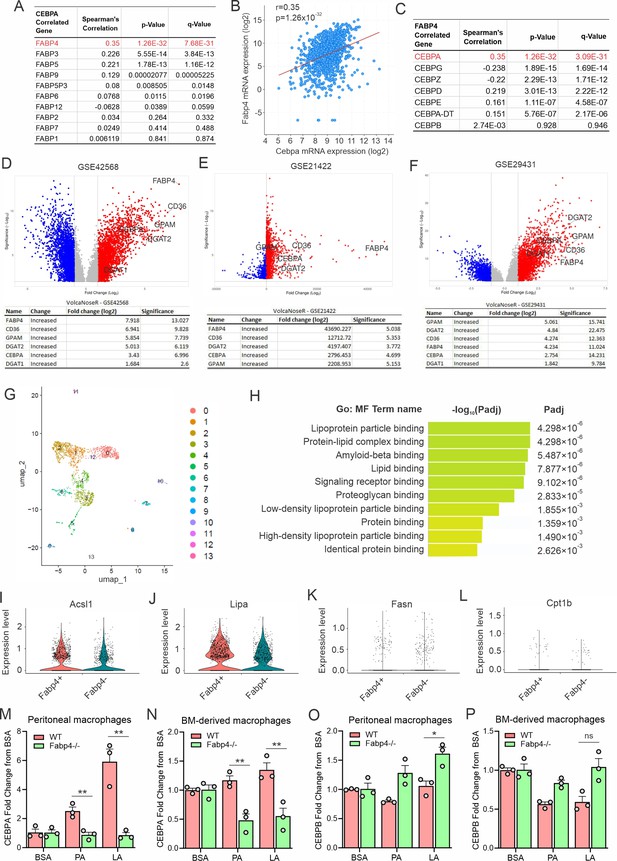
FABP4 mediates LA-induced CEBPα, but not CEBPβ, activation.
(A, B) Spearman’s correlation analysis of CEBPα with FABP family members, especially FABP4 with CEBPα (B), using cBioportal breast cancer TCGA database. (C) Spearman’s correlation analysis of FABP4 with CEBP family members using cBioportal breast cancer TCGA database. (D–F) Differential gene expression analysis of FABP4, GPAM, DGAT1, DGAT2, CEBPA, and CD36 in published breast cancer microarray databases, including GSE42568 (D), GSE21422 (E) and GSE29431 (F). (G) UMAP of different subsets of splenic macrophages using single-cell RNA sequence analysis. (H) Molecular function pathway analysis of the integrated conserved genes in the FABP4-positive clusters by g:Profiler. (I–L) Violin plots showing relative expression levels of genes, including Acs1(I), Lipa (J) and Fasn (K) and Cpt1b (L) between FABP4 +vs FABP4- macrophages in the splenic macrophage single cell RNA sequencing analysis. (M) Measurement of CEBPA gene levels in WT and FABP4-/- peritoneal macrophages treated with BSA, PA, or LA, respectively, for 4 hr. (N) Measurement of CEBPA gene levels in WT and FABP4-/- bone-marrow-derived macrophages treated with BSA, PA, or LA, respectively, for 4 hr. (O) Measurement of CEBPB gene levels in WT and FABP4-/- peritoneal macrophages treated with BSA, PA, or LA, respectively, for 4 hr. (P) Measurement of CEBPB gene levels in WT and FABP4-/- bone-marrow-derived macrophages treated with BSA, PA, or LA, respectively, for 4 hr. Data are shown as mean ± SD in panels I-P (p* p≤0.05, ** p≤0.01, ns, non-significant, unpaired Student t test).
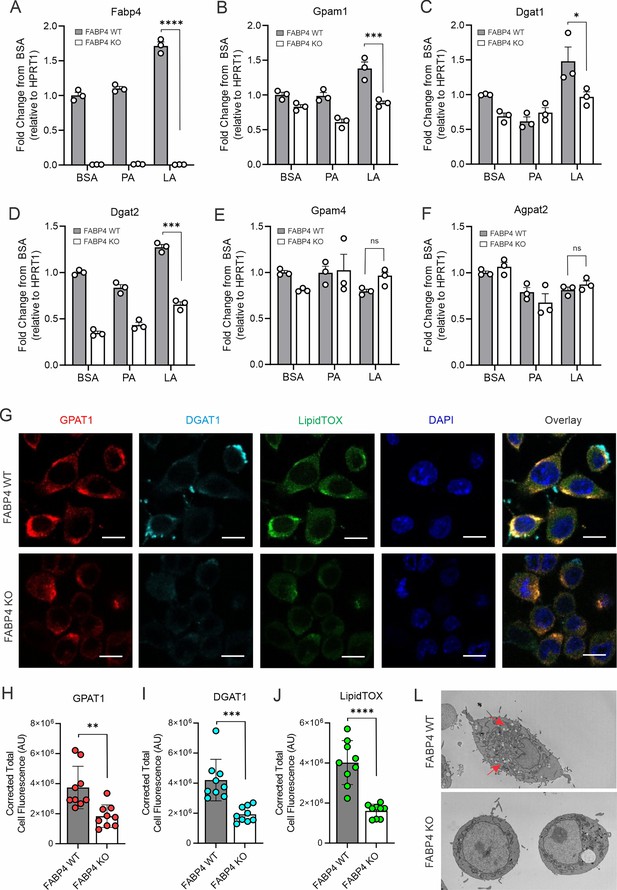
FABP4 deficiency reduces LA-induced lipid accumulation in macrophages.
(A–F) real-time PCR analysis of FABP4 (A) and genes encoding key enzymes for triglycerol biosynthesis, including Gpam1 (B), Dgat1 (C), Dgat2 (D), Gpam4 (E), Agpat2 (F) in WT and FABP4 KO macrophages treated with BSA, PA, or LA (400 μM). (G) Confocal analysis of protein expression of GPAT1 (red), DGAT1 (cyan) and lipid accumulation (LipidTOX staining, green) in LA-treated WT and FABP4 KO macrophages (bar, 10 μM). (H–J) Expression levels of GPAT1 (H), DGAT1 (I) and LipidTOX (J) as indicated in panel G were quantified by Image J. (L) Flow cytometric analysis of neutral lipid accumulation as shown by BODIPY staining in WT and FABP4 KO macrophages treated with BSA or LA. (M) Transmission electron microscope showing lipid droplet staining in WT and FABP4 KO macrophages treated with LA. Data are shown as mean ± SD in panels A-F, H-L (* p≤0.01, p** p≤0.01, *** p≤0.001, ns, non-significant as compared to the control BSA group or FABP4-/- group, unpaired Student t test).
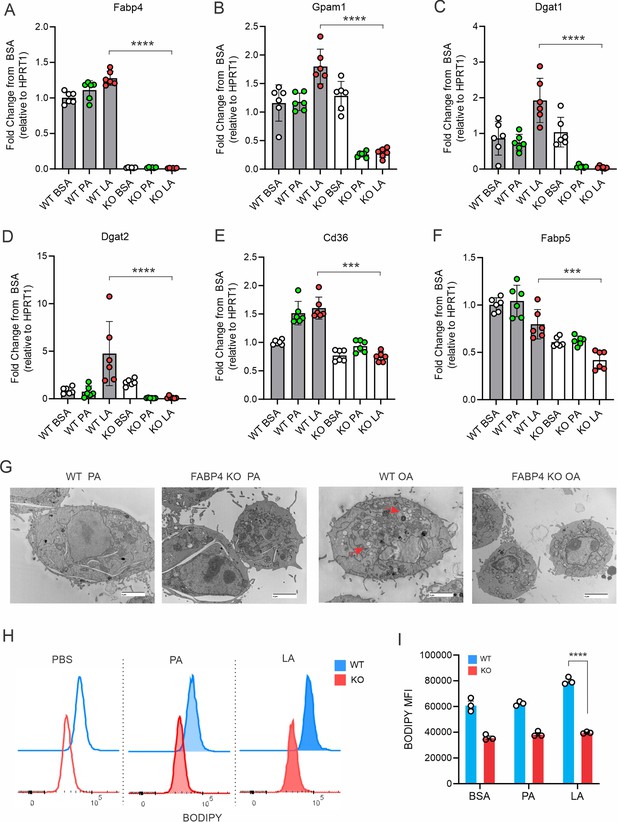
Deficiency of FABP4 reduces lipid droplet formation in macrophages.
(A–F) Real-time PCR analysis of FABP4 (A) and genes encoding key enzymes for triglycerol biosynthesis, including Gpam1 (B), Dgat1 (C), Dgat2 (D), Cd36 (E), Fabp5 (F) in WT and FABP4 KO peritoneal macrophages treated with BSA, PA, and LA (400 μM). (G) Transmission electron microscope showing lipid droplet formation (red arrows) in WT and KO macrophages treated with either PA or OA for 4 hr. (H, I) Flow cytometric analysis of BODIPY staining (G) and mean fluorescent intensity (MFI) in peritoneal macrophages treated with BSA, PA, or LA (400 μM). Data are shown as mean ± SD (*** p≤0.001, *** p≤0.001, unpaired Student t test).
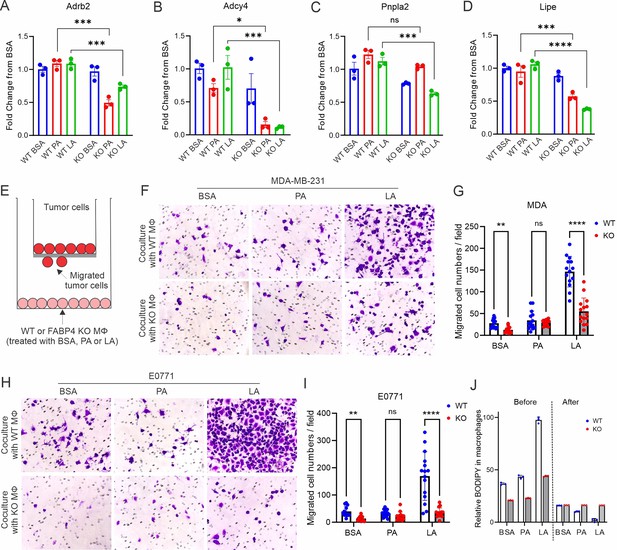
FAPB4 expression in macrophages promotes lipolysis and breast cancer cell migration.
(A–D) Realtime PCR analysis of expression of Adrb2 (A), Adcy4 (B), Pnpla2 (C) and Lipe (D) in FABP4 WT and KO macrophages treated with BSA, PA, or LA (400 μM). (E) Transwell measurement of migration of breast cancer cells cocultured with FA- or BSA-treated FABP4 WT or KO macrophages (Mφ). (F–I) FABP4 WT or KO macrophages were treated with 100 μM BSA, PA, or LA for 4 hr. Fatty acids in the culture medium were washed away with FBS-free RPMI-1640. Breast cancer cells were added to a transwell and cocultured with these different FA-or BSA-treated FABP4 WT or KO macrophages for 24 hr. The migrated tumor cells were stained and quantified. Migrated MDA-MB-231 cells were shown in panels F and G. Migrated E0771 cells were shown in panels H and I. (J) FABP4 WT and KO macrophages were treated with indicated FAs or BSA for 4 hr. Flow cytometric staining of BODIPY levels in WT and KO macrophages before and after coculture with E0771 tumor cells for 24 hr. Data are shown as mean ± SD in panels A-D, G, I, J (* p≤0.01, p** p≤0.01, *** p≤0.001, ns, non-significant as compared to the control group or FABP4 KO group, unpaired Student t test).
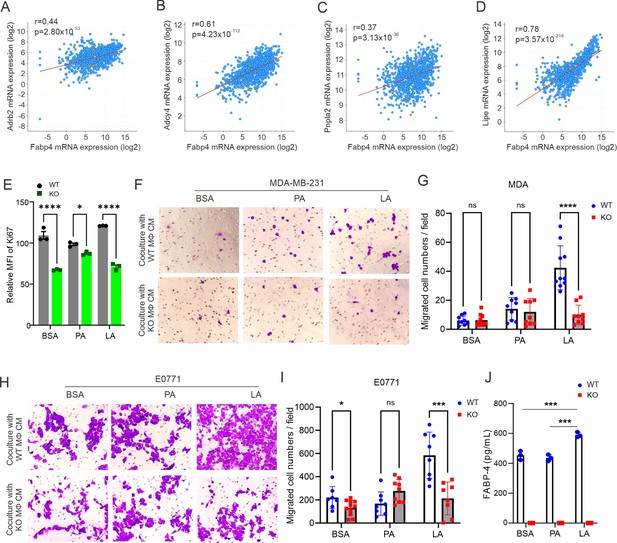
FABP4 promotes lipolysis and tumor migration.
(A–D) Spearman’s correlation analysis between FABP4 and key genes mediating lipolysis, including Adrb2 (A), Adcy4 (B), Pnpla2 (C), and Lipe (D) using the cBioportal breast cancer TCGA database. (E) Measurement of cell proliferation marker Ki67 in E0771 tumor cells after coculture with FA-treated FABP4 WT or KO macrophages for 24 hr. (F, G) After treatment with BSA, PA, or LA (400 μM) for 4 hr, FABP4 WT and KO macrophages were cultured in FBS-free RPMI-1640 for 24 hr. The cultural conditional medium was collected for tumor migration assays. MDA-MB-231 migration and quantification are shown in panels F and G, respectively. (H, I) E0771 tumor migration assays were performed using the conditional medium collected as above plus 1% FBS. Tumor migration and quantification are shown in panels H and I, respectively. (J) ELSIA measurement of FABP4 levels in macrophage/tumor coculture medium. Data are shown as mean ± SD in panels G, I, and J (p* p≤0.05, *** p≤0.001, ns, non-significant, unpaired Student t test).
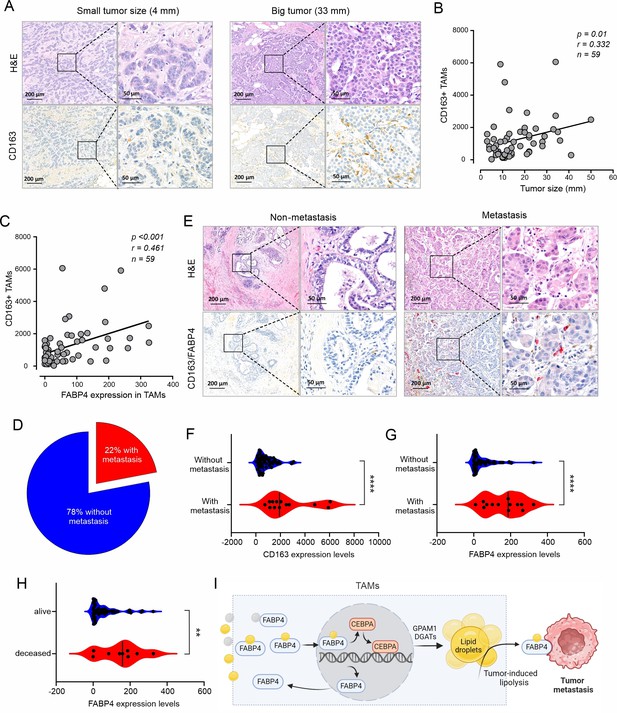
High expression of FABP4 in TAMs is associated with more metastasis of breast cancer.
(A) Comparison of H&E and CD163 staining (brown) between an example of small and large breast cancer tumors in breast cancer patients. (B) Spearman correlation analysis between breast cancer tumor size and CD163 +TAM staining. (C) Expression of FABP4 and CD163 was highly correlated as analyzed by the Spearman correlation analysis in breast cancer tissues. (D) Pie chart showing the percentage of breast cancer patients with or without metastasis. (E) Analysis of the staining of H&E, CD163 (brown), FABP4 (red) in primary breast tumors of patients with and without metastasis. (F) Analysis of CD163 expression levels between primary breast tumors of patients with and without metastases. (G) Analysis of FABP4 expression levels between primary breast tumors of patients with and without metastases. (H) Analysis of FABP4 expression levels between alive and deceased breast cancer patients. (I) Scheme of how FABP4 mediates unsaturated FA (yellow)-induced lipid storage and lipolysis in TAMs. When TAMs are exposed to dietary saturated (gray) or unsaturated (yellow) FAs, unsaturated FAs, but not saturated ones, induce FABP4 nuclear translocation and upregulate FABP4 and CEBPA-mediated transactivation of GPAM1 and DGATs, promoting lipid storage as lipid droplets. Once tumor-induced lipolysis occurs, FABP4/unsaturated FAs are secreted from TAMs to induce tumor migration and metastasis. Data are shown as mean ± SD in panel F-H (p** p0.01, *** p≤0.001, *** p≤0.001, unpaired Student t test).
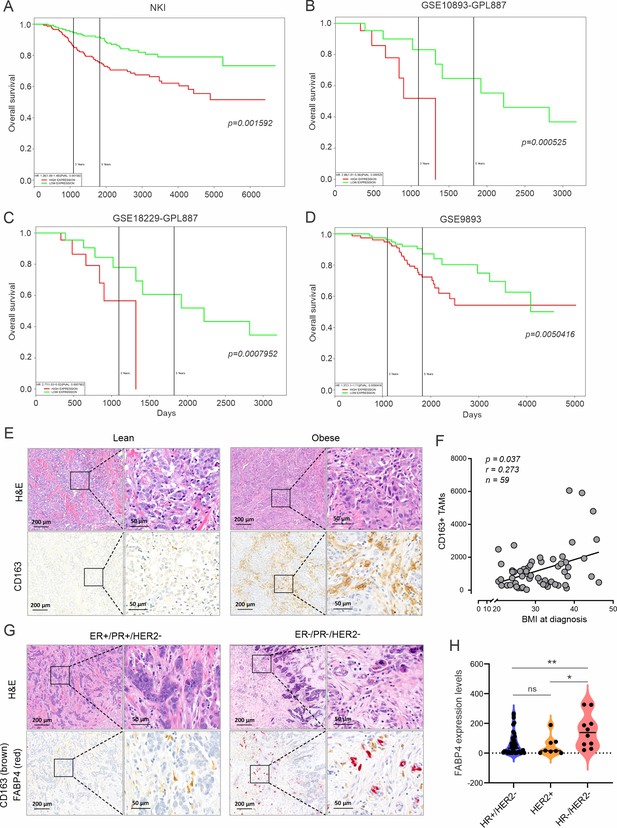
Association of TAMs with obesity and survival of breast cancer patients.
(A–D) PROGgene analysis of the association between CD163 expression levels and breast cancer survival using various publicly accessible breast cancer databases, including NKI (A), GSE10893 (B), GSE18229 (C) and GSE9893 (D). (E) Comparison of H&E and CD163 staining (brown) between lean and obese breast cancer tumors in breast cancer patients. (F) Spearman’s correlation analysis between CD163 expression in breast cancer tissues and BMI at diagnosis of breast cancer patients. (G) H&E and immunohistochemistry staining of FABP4 (red) in ER +and triple negative breast cancer tissues. (H) Analysis of FABP4 expression in hormone receptor+/HER2-, HER2 +and triple negative breast cancers. Data are shown as mean ± SD in panel H (p* p≤0.05, ** p≤0.01, ns, non-significant, unpaired Student t test).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | PE/Cyanine7 rat anti-mouse F4/80 monoclonal | BioLegend | Cat#123114 | 1:500 |
| Antibody | Brilliant Violet 711 Armenian Hamster anti-mouse CD11c monoclonal | BioLegend | Cat#117349 | 1:500 |
| Antibody | Brilliant Violet 605 rat anti-mouse I-A/I-E monoclonal | BioLegend | Cat#107639 | 1:500 |
| Antibody | BUV737 rat anti-mouse CD11b monoclonal | BD Biosciences | Cat#612800 | 1:500 |
| Antibody | BUV563 rat anti-mouse CD45 monoclonal | BD Biosciences | Cat#752412 | 1:500 |
| Antibody | Goat anti-human FABP4polyclonal | R&D Systems | Cat#AF3150 | 1:300 |
| Antibody | Goat anti-mouseFABP4 polyclonal | R&D Systems | Cat#AF1443 | 1:300 |
| Antibody | Rabbit anti-human/mouse GPAM polyclonal | Invitrogen | Cat#PA5-20524 | 1:300 |
| Antibody | Goat anti-human/mouse DGAT1 polyclonal | Sigma-Aldrich | Cat#SAB2500307 | 1:300 |
| Antibody | Rabbit anti-human/mouse C/EBPα polyclonal | Cell Signaling Technology | Cat#2295 | 1:300 |
| Antibody | Mouse anti-human C/EBPβ monoclonal | DSHB | Cat#PCRP-CEBPB-3D10 | 1:300 |
| Antibody | Rabbit anti-CD163 human/mouse monoclonal | Abcam | Cat#EPR19518 | 1:300 |
| Antibody | Alexa Fluor 594-conjugated AffiniPure mouse anti-goat IgG polyclonal | Jackson ImmunoResearch | Cat#205-605-108 | 1:2000 |
| Antibody | Alexa Fluor 647-conjugated AffiniPure donkey anti-goat IgG polyclonal | Jackson ImmunoResearch | Cat#705-586-147 | 1:2000 |
| Antibody | Alexa Fluor Plus 488 donkey anti-goat IgG polyclonal | Invitrogen | Cat#A32814 | 1:2000 |
| Antibody | Alexa Fluor 647-conjugated AffiniPure Goat anti-rabbit IgG polyclonal | Jackson ImmunoResearch | Cat#111-605-003 | 1:2000 |
| Antibody | Alexa Fluor 546 goat anti-rabbit IgG polyclonal | Invitrogen | Cat#A11035 | 1:2000 |
| Antibody | Alexa Fluor 488 goat anti-mouse IgG polyclonal | Invitrogen | Cat#A11029 | 1:2000 |
| Antibody | Alexa Fluor 546 goat anti-mouse IgG polyclonal | Invitrogen | Cat#A11030 | 1:2000 |
| Chemical compound, drug | Ghost Dye Violet 510 Viability Dye | Tonbo Biosciences | Cat#13–0870 T100 | 1:200 |
| Chemical compound, drug | Bodipy 493/503 | Molecular Probes | Cat#D3922 | Final concentration: 5 µM |
| Chemical compound, drug | LysoTracker Blue DND-22 | Invitrogen | Cat#L7525 | Final concentration: 50 nM |
| Chemical compound, drug | Cell Navigator Live Cell Endoplasmic Reticulum Staining Kit Red | ATT Bioquest | Cat#22636 | 500 × |
| Chemical compound, drug | MitoSpy NIR DiIC1(5) | BioLegend | Cat#424807 | Final concentration: 10 nM |
| Chemical compound, drug | HCS LipidTOX Green Neutral Lipid Stain | Invitrogen | Cat#H34475 | 1000 × |
| Chemical compound, drug | Hoechst 33342 Solution | Invitrogen | Cat#62249 | Final concentration: 0.1 µg/ml |
| Chemical compound, drug | DAPI solution (1 mg/mL) | Invitrogen | Cat#62248 | 1:1000 |
| Chemical compound, drug | Oil Red O powder | Alfa Aesar | Cat#A12989.14 | Final concentration: 1.8 mg/ml |
| Chemical compound, drug | Phosphate Buffered Saline pH 7.4 (PBS) | Gibco | Cat#10010–023 | |
| Chemical compound, drug | Paraformaldehyde Solution 4% in PBS | Thermo Fisher Scientific | Cat#J19943.K2 | |
| Chemical compound, drug | Neutral Buffered Formalin Solution 10% in PBS | VWR | Cat#10790–714 | |
| Chemical compound, drug | Triton X-100 | Sigma-Aldrich | Cat#X100-500ML | Final concentration: 0.2% |
| Chemical compound, drug | ProLong Diamond Antifade Mountant | Invitrogen | Cat#P36961 | |
| Chemical compound, drug | RPMI Medium 1640 | Gibco | Cat#11875–093 | |
| Chemical compound, drug | Seahorse XF RPMI Meduim pH7.4 | Agilent Technologies | Cat#103576–100 | |
| Chemical compound, drug | Seahorse XF calibrant solution | Agilent Technologies | Cat#100840–000 | |
| Chemical compound, drug | Penicillin-Streptomycin | Thermo Fisher Scientific | Cat#15140122 | 100 × |
| Chemical compound, drug | Fetal Bovine Serum | R&D Systems | Cat#S11550 | |
| Chemical compound, drug | Goat Serum | Thermo Fisher Scientific | Cat#31873 | |
| Chemical compound, drug | Palmitate | Nu-Chek Prep, Inc | S-1109 | 5 mM in stock |
| Chemical compound, drug | Stearate | Nu-Chek Prep, Inc | S-1111 | 5 mM in stock |
| Chemical compound, drug | Oleate | Nu-Chek Prep, Inc | S-1120 | 5 mM in stock |
| Chemical compound, drug | Linoleate | Nu-Chek Prep, Inc | S-1127 | 5 mM in stock |
| Chemical compound, drug | Bovine Serum Albumin | Proliant | Cat# 69760 | 5 mM in stock |
| Chemical compound, drug | Poly-L-lysine | Sigma-Aldrich | Cat#P9155 | 0.5 ml of a 0.1 mg/ml solution to coat 25 cm2 |
| Chemical compound, drug | jetPRIME | Sartorius | Cat#101000046 | |
| Chemical compound, drug | Power SYBR Green PCR Master Mix | Applied Biosystems | Cat#4368708 | |
| Chemical compound, drug | RBC Lysis Buffer | Tonbo Biosciences | Cat#TNB-4300-L100 | |
| Commercial assay, kit | Chromium NextGEM Chip G Single Cell Kit | 10 X Genomics | Cat#1000127 | |
| Commercial assay, kit | Dual Index Kit TT Set A | 10 X Genomics | Cat#1000215 | |
| Commercial assay, kit | Chromium NextGEM Single Cell 3’Kit v3.1 | 10 X Genomics | Cat#1000269 | |
| Commercial assay, kit | Magnetic Separator | 10 X Genomics | Cat#120250 | |
| Commercial assay, kit | Non-acetylated BSA | 10 X Genomics | Cat#B9000S | |
| Commercial assay, kit | Seahorse XF Cell Mito Stress Test Kit | Agilent Technologies | Cat#103015–100 | |
| Commercial assay, kit | Seahorse XF96 Cell Culture Plate | Agilent Technologies | Cat#101085–004 | |
| Commercial assay, kit | Seahorse XFe96 Extracellular flux assay kits | Agilent Technologies | Cat#102601–100 | |
| Commercial assay, kit | PureLink RNA mini kit | Invitrogen | Cat#12183025 | |
| Commercial assay, kit | QuantiTect reverse transcription kit | Qiagen | Cat#205314 | |
| Commercial assay, kit | Zombie Violet Fixable Viability Kit | Biolegend | Cat#423113 | 1:1000 |
| Commercial assay, kit | ImmPRESS HRP Horse Anti-Rabbit IgG Polymer Detection Kit, Peroxidase | Vector | Cat#MP-7401–50 | |
| Commercial assay, kit | BLOXALL Endogenous Blocking Solution, Peroxidase and Alkaline Phosphatase | Vector | SP-6000–100 | |
| Commercial assay, kit | CircuLex Mouse FABP4/A-FABP ELISA Kit | CircuLex/MBL | Cat#CY-8077 | |
| Sequence-based reagent | DsiRNA for mouse C/EBPα | IDT | Cat#mm.Ri.Cebpa.13.1 | Transfection at final concentration of 50 nM |
| Sequence-based reagent | DsiRNA for mouse C/EBPβ | IDT | Cat#mm.Ri.Cebpb.13.1 | Transfection at final concentration of 50 nM |
| Peptide, recombinant protein | Recombinant Mouse M-CSF | BioLegend | Cat#576406 | Final concentration: 30 ng/ml |
| Recombinant DNA reagent | Microarray data (Breast Cancer Gene Expression Analysis) | PMID:23740839 | GSE42568 | |
| Recombinant DNA reagent | Microarray data (Expression profiling of human DCIS and invasive ductal breast carcinoma) | PMID:21468687 | GSE21422 | |
| Recombinant DNA reagent | Microarray data (Identifying breast cancer biomarkers) | PMID:141503 | GSE29431 | |
| Recombinant DNA reagent | NKI | PROGeneV2 | https://bioconductor.org/packages/breastCancerNKI | |
| Recombinant DNA reagent | Microarray data | PROGeneV2 | GSE10893-GPL887 | |
| Recombinant DNA reagent | Microarray data | PROGeneV2 | GSE18229-GPL887 | |
| Recombinant DNA reagent | Microarray data | PROGeneV2 | GSE9893 | |
| Cell line (mouse) | Bone Marrow Derived Macrophage-J2 Immortalized | This Paper | N/A | See the Methods details – Cell lines |
| Cell line (mouse) | FABP4-/- Bone Marrow Derived Macrophage-J2 Immortalized | This Paper | N/A | See the Methods details – Cell lines |
| Cell line (mouse) | BMM | This paper | N/A | See the Methods details – Primary cells |
| Cell line (mouse) | Peritoneal Macrophage | This paper | N/A | See the Experimental model and study participant details – Mice |
| Cell line (mouse) | E0771 | ATCC | Cat#CRL-3461 | |
| Cell line (human) | MDA-MB 231 | ATCC | Cat#HTB-26 | |
| Strain, strain background | Mouse:C57Bl/6 J | Jackson Laboratory | JAX 000664 | |
| Strain, strain background | Mouse:FABP4-/- | This Paper | See the Experimental model and study participant details – Mice | |
| Sequence-based reagent | Real-time PCR Primers | Supplementary file 3 | N/A | |
| Software and algorithms | FlowJo v10 | BD Biosciences | https://www.flowjo.com/ | |
| Software, algorithm | GraphPad Prism 8 | GraphPad Software | https://www.graphpad.com/ | |
| Software, algorithm | ImageJ (Fiji edition) | NIH | https://imagej.net/software/fiji/ | |
| Software, algorithm | Design and Analysis Software V2.3 | Thermo Fisher | http://www.thermofisher.com/us/en/home/global/forms/life-science/quantstudio-6-7-pro-software.html | |
| Software, algorithm | Seahorse Wave | Agilent Technologies | https://www.agilent.com/zh-cn/product/cell-analysis/real-time-cell-metabolic-analysis/xf-software/seahorse-wave-desktop-software-740897 | |
| Software, algorithm | Web-based Gene Set Analysis Toolkit | WebGestalt | https://www.webgestalt.org/ | |
| Software, algorithm | VolcaNoseR2 | VolcaNoseR2 | https://huygens.science.uva.nl/VolcaNoseR2/ | |
| Software, algorithm | g:Profiler | ELIXIR | https://biit.cs.ut.ee/gprofiler/gost | |
| Software, algorithm | CiiiDER | CiiiDER | The program and documentation are available from https://www.ciiider.org/ and the source code is available at https://gitlab.erc.monash.edu.au/ciiid/ciiider (Gearing, 2020). | Refer to https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0215495 |
| Software, algorithm | CoreIDRAW X7 | CoreIDRAW | https://www.coreldraw.com/en/ | |
| Software, algorithm | Halo Software | Indica Labs | V3.6.4134.263 | |
| Other | Sterile 24 well cell culture plate | Corning | Cat#3526 | See the Method details – Migration assay |
| Other | Sterile 100 mm cell culture dish | Greiner Bio-One | Cat#664160 | See the Method details – Primary cells |
| Other | Sterile 25Gx1” Needle | Becton, Diskinson and Company | Cat#305125 | See the Method details – Primary cells |
| Other | Sterile 10 mL syringe | Becton, Diskinson and Company | Cat#302995 | See the Method details – Primary cells |
| Other | Sterile 15 mL conical-bottom Centrifuge Tube | Avantor | Cat#525–1068 | See the Method details – Primary cells |
| Other | 40 µm cell strainer | VWR | Cat#BX15-1040 | See the Method details – Primary cells |
| Other | Transwell inserts 8.0 µm pore size | Falcon | Cat#353097 | See the Method details – Migration assay |
| Other | 5 mL Polystyrene round-bottom tube | Falcon | Cat#352058 | See the Method details – Flow cytometry |
| Other | MicroAmp Optical 384-well Reaction Plate | Applied Biosystems | Cat#4309849 | See the Method details – Quantitative RT-PCR |
| Other | QuantStudio 7 Flex Real-Time PCR system | Applied Biosystems | Cat#4485701 | See the Method details – Quantitative RT-PCR |
| Other | Seahorse XFe96 extracellular flux analyzer | Seahorse Biosciences | See the Method details – Seahorse cell mito stress | |
| Other | Cytek Aurora | CytekBio | See the Method details – Flow cytometry | |
| Other | Cytek Amnis ImageStream MkII Imaging Flow Cytometer | CytekBio | See Figure 1F | |
| Other | Synergy LX Multi-Mode Microplate Reader | BioTek | Cat#SLXFATS-SN | See the Method details – ELISA for FABP4 analysis |
| Other | Echo Revolution 2 microscope | ECHO | See Figure 6F and 6 H | |
| Other | Zeiss LSM880-airyscan | Zeiss | See Figure 2I, Figure 3B and L, Figure 4K and L, and Figure 5G | |
| Other | Electron Microscope | Hitachi | HT7800 | See the Method details – Transmission electron microscopy |
| Other | Slide Scanner | Leica | Aperio GT 450 | See Figure 7 |
Additional files
-
Supplementary file 1
Patient information.
- https://cdn.elifesciences.org/articles/101221/elife-101221-supp1-v1.xlsx
-
Supplementary file 2
Summary statistics for categorical variables and their association with outcome variables.
- https://cdn.elifesciences.org/articles/101221/elife-101221-supp2-v1.docx
-
Supplementary file 3
Realtime PCR primer sequences.
- https://cdn.elifesciences.org/articles/101221/elife-101221-supp3-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/101221/elife-101221-mdarchecklist1-v1.pdf






