Identification of the trail-following pheromone receptor in termites
Figures
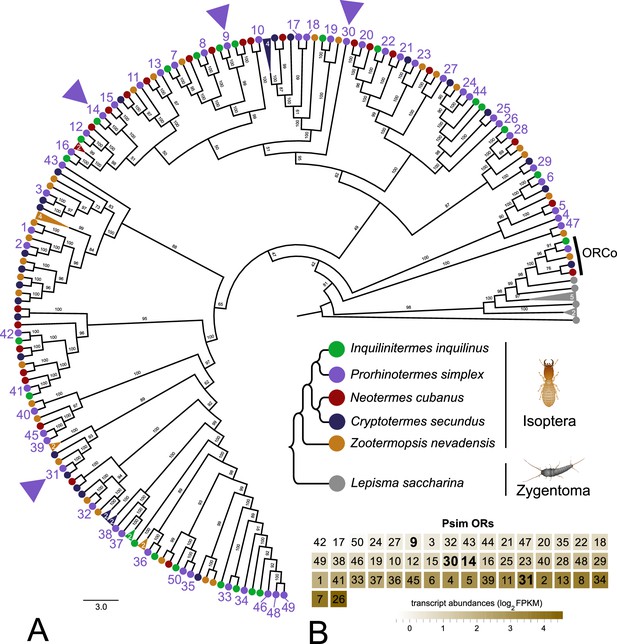
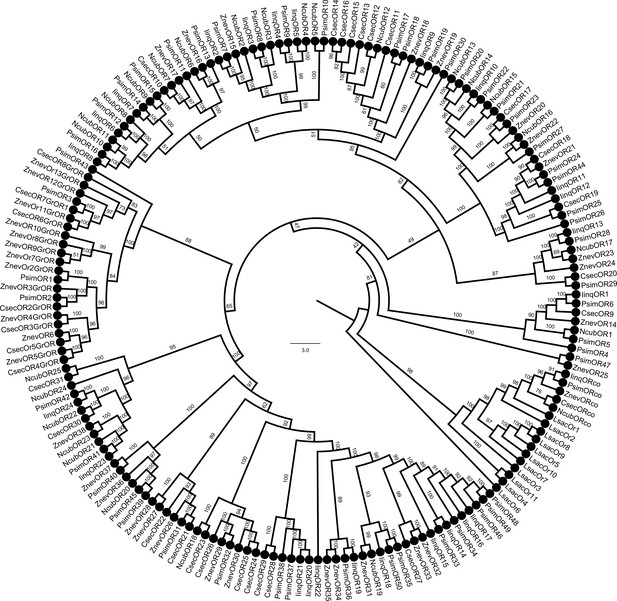
Phylogenetic reconstruction of termite odorant receptors (ORs) and their transcript abundances in Prorhinotermes simplex workers.
(A) Phylogenetic tree is based on 182 protein sequences from five species of termites and the bristletail Lepisma saccharina as a basal insect outgroup, and also includes the sequences of ORCo. The topology and branching supports were inferred using the IQ-TREE maximum likelihood algorithm with the JTT+F+R8 model and supported by 10,000 iterations of ultrafast bootstrap approximation. Protein sequences of termite ORs can be found under the same labeling in Johny et al., 2023. L. saccharina sequences are listed in Thoma et al., 2019. Arrowheads highlight the four ORs from P. simplex selected for functional characterization. A fully annotated version of the tree is provided as Figure 1—figure supplement 1. (B) Heatmap shows the transcript abundances of 50 ORs identified in the RNAseq data from P. simplex worker antennae available in NCBI SRA archive under accession SRX17749141.
Fully annotated version of the phylogenetic tree of termite odorant receptors (ORs) shown in Figure 1.
Protein sequences of termite ORs can be found under the same labeling in Johny et al., 2023. Lepisma saccharina sequences used as basal insect outgroup are listed in Thoma et al., 2019. The topology and branching supports were inferred using the IQ-TREE maximum likelihood algorithm with the JTT+F+R8 model and supported by 10,000 iterations of ultrafast bootstrap approximation.
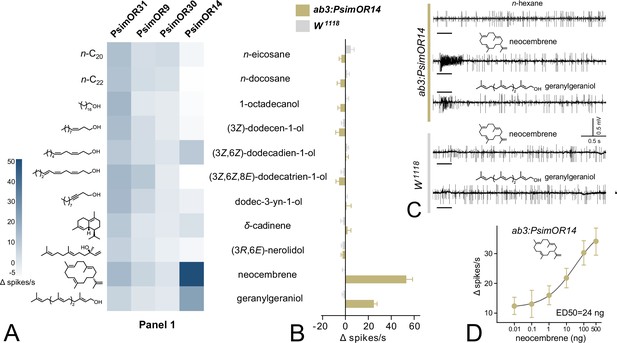
Single-sensillum recording (SSR) responses of transgenic Drosophila melanogaster ab3 sensillum expressing PsimOR9, 14, 30, and 31 to the initial screening of 11 volatiles with biological relevance for termites.
(A) Heatmap showing the average responses of the four odorant receptors (ORs) as Δ spikes/s from 3 to 6 independent replicates. (B) Comparison of SSR responses of transgenic D. melanogaster ab3A neurons expressing PsimOR14 (ab3A:PsimOR14) and W1118 D. melanogaster. The bars show the average Δ spikes/s values from five independent replicates ± SEM. (C) Characteristic SSR traces of ab3A:PsimOR14 and W1118 flies for 1 µg dose of neocembrene and geranylgeraniol. (D) Dose–response curve of ab3A:PsimOR14 SSR responses to neocembrene. The graph shows average Δ spikes/s values ± SEM based on nine replicates (8 in case of 100 ng and 4 in case of 500 ng stimulations). The curve fit and ED50 value were calculated using log(agonist) versus response non-linear algorithm with least square fit method and the constraint of minimal response >0. The crossing scheme for transgenic fly generation is shown in Figure 2—figure supplement 1, the raw data for all graphs is provided in Figure 2—source data 1–6.
-
Figure 2—source data 1
Single-sensillum recording (SSR) responses to Panel 1 for PsimOR9.
- https://cdn.elifesciences.org/articles/101814/elife-101814-fig2-data1-v1.xlsx
-
Figure 2—source data 2
Single-sensillum recording (SSR) responses to Panel 1 for PsimOR14.
- https://cdn.elifesciences.org/articles/101814/elife-101814-fig2-data2-v1.xlsx
-
Figure 2—source data 3
Single-sensillum recording (SSR) responses to Panel 1 for PsimOR30.
- https://cdn.elifesciences.org/articles/101814/elife-101814-fig2-data3-v1.xlsx
-
Figure 2—source data 4
Single-sensillum recording (SSR) responses to Panel 1 for PsimOR31.
- https://cdn.elifesciences.org/articles/101814/elife-101814-fig2-data4-v1.xlsx
-
Figure 2—source data 5
Single-sensillum recording (SSR) responses to Panel 1 for ab3:PsimOR14 versus W1118.
- https://cdn.elifesciences.org/articles/101814/elife-101814-fig2-data5-v1.xlsx
-
Figure 2—source data 6
Single-sensillum recording (SSR) dose–response data for neocembrene and PsimOR14 fly line.
- https://cdn.elifesciences.org/articles/101814/elife-101814-fig2-data6-v1.xlsx
Crossing scheme of termite odorant receptors (ORs) heterologous expression using Drosophila melanogaster empty neurons in ab3 sensilla.
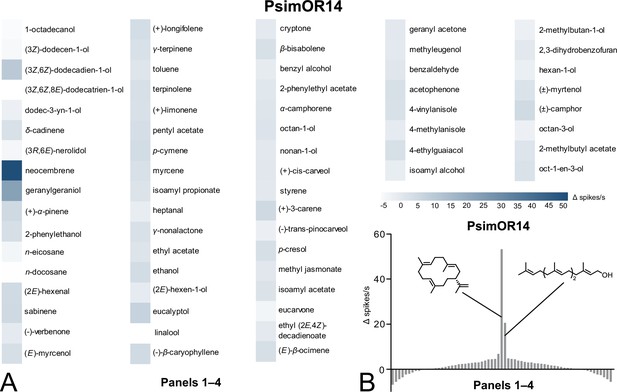
Single-sensillum recording (SSR) responses of transgenic D. melanogaster ab3 sensillum expressing PsimOR14 to the complete set of 67 compounds (Panels 1–4).
(A) Heatmap showing the average responses as Δ spikes/s from 3 to 6 independent replicates. (B) Tuning curve of PsimOR14 for the 67 compounds contained in Panels 1–4. The raw data for both graphs is provided in Figure 3—source data 1–3. Origin and purity of the tested chemicals are provided in Figure 3—source data 4.
-
Figure 3—source data 1
Single-sensillum recording (SSR) responses to Panel 2 for PsimOR14.
- https://cdn.elifesciences.org/articles/101814/elife-101814-fig3-data1-v1.xlsx
-
Figure 3—source data 2
Single-sensillum recording (SSR) responses to Panel 3 for PsimOR14.
- https://cdn.elifesciences.org/articles/101814/elife-101814-fig3-data2-v1.xlsx
-
Figure 3—source data 3
Single-sensillum recording (SSR) responses to Panel 4 for PsimOR14.
- https://cdn.elifesciences.org/articles/101814/elife-101814-fig3-data3-v1.xlsx
-
Figure 3—source data 4
Origin and purity of the tested chemicals.
- https://cdn.elifesciences.org/articles/101814/elife-101814-fig3-data4-v1.xlsx
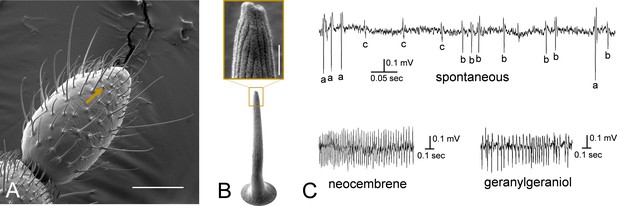
Neocembrene-responding sensillum in P. simplex workers.
(A) Scanning electron microscopy (SEM) photograph of the last flagellomere of P. simplex worker. Arrow shows a small multiporous grooved sensillum responding to neocembrene and geranylgeraniol. Scale bar represents 50 µm. (B) High-resolution SEM (HR-SEM) view on the neocembrene-responding sensillum. Scale bar in the inset represents 500 nm. (C) Detailed view on single-sensillum recording (SSR) traces recorded from the neocembrene-responding sensillum during spontaneous firing, and upon stimulation with neocembrene and geranylgeraniol.
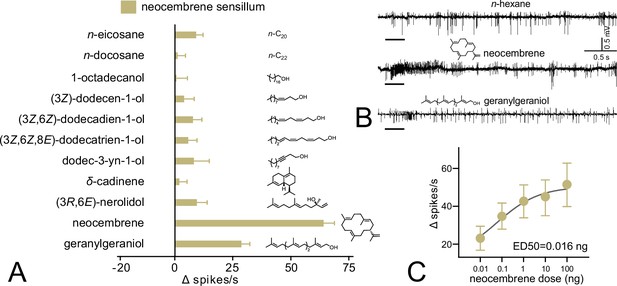
Single-sensillum recording (SSR) responses of the neocembrene-responding sensillum on the last flagellomere of P. simplex worker.
(A) SSR responses to Panel 1. The bars show the average Δ spikes/s values from 8 to 17 replicates ± SEM. The raw data is provided in Figure 5—source data 1. (B) Characteristic SSR traces of the neocembrene-detecting sensillum for neocembrene and geranylgeraniol. (C) Dose–response curve of the SSR responses to neocembrene by the neocembrene-responding sensillum. The graph shows average Δ spikes/s values ± SEM based on 9–11 replicates. The curve fit and ED50 value were calculated using log(agonist) versus response non-linear algorithm with least square fit method and the constraint of minimal response >0. The raw data is provided in Figure 5—source data 2.
-
Figure 5—source data 1
Single-sensillum recording (SSR) responses to Panel 1 by neocembrene sensillum in P. simplex workers.
- https://cdn.elifesciences.org/articles/101814/elife-101814-fig5-data1-v1.xlsx
-
Figure 5—source data 2
Single-sensillum recording (SSR) dose–response data for neocembrene and P. simplex neocembrene sensillum.
- https://cdn.elifesciences.org/articles/101814/elife-101814-fig5-data2-v1.xlsx
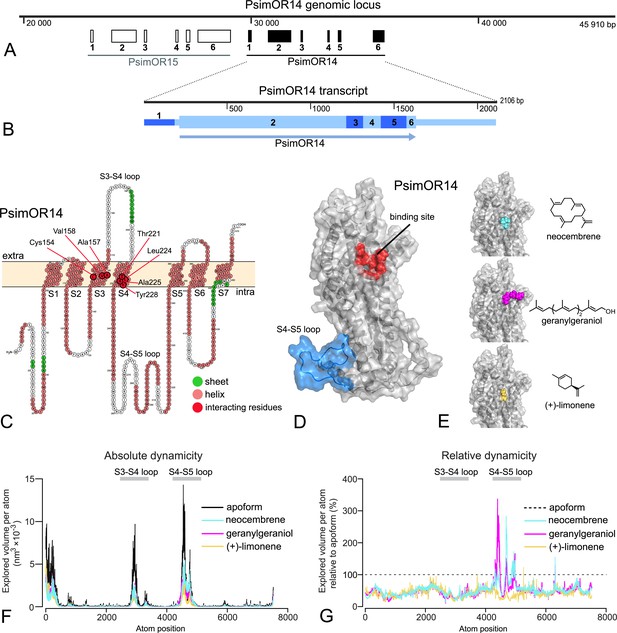
PsimOR14 gene, transcript, and protein structures, docking, and molecular dynamics (MD) simulations.
(A) Genomic locus containing PsimOR14 and PsimOR15. PsimOR14 gene consists of one non-coding and five protein-coding exons. (B) PsimOR14 transcript with six exons, showing the protein-coding (higher boxes) and untranslated regions (lower boxes), and open reading frame (ORF; arrow). (C) Transmembrane architecture of PsimOR14. In red are shown seven residues interacting with neocembrene. Light blue ellipse shows the intracellular loop the most impacted by ligand binding. (D) Modeled apoform of PsimOR14. Red region denotes the binding site identified via docking, light blue region represents the intracellular S4–S5 loop. (E) Holoforms of PsimOR14 with three docked ligands. (F) Absolute PsimOR4 dynamicity expressed as average volumes explored by atoms per simulation step in PsimOR14 apoform and upon binding the three studied ligands. (G) Relative PsimOR14 dynamicity expressed as average explored atom volumes upon ligand binding relative to the volumes in PsimOR14 apoform. Nucleotide and protein sequences of PsimOR14 are provided under NCBI entry OR921181 and as Figure 6—source data 1. Interacting residues and their binding energies to the ligands are listed in Figure 6—source data 2. The raw data for explored atomic volumes are provided in Figure 6—source data 3.
-
Figure 6—source data 1
Nucleotide and amino acid sequences of PsimOR14.
- https://cdn.elifesciences.org/articles/101814/elife-101814-fig6-data1-v1.docx
-
Figure 6—source data 2
Residues interacting with the docked ligands and their binding energies.
- https://cdn.elifesciences.org/articles/101814/elife-101814-fig6-data2-v1.xlsx
-
Figure 6—source data 3
Explored atomic volumes of PsimOR14 apoform and upon binding of ligands, inferred from molecular dynamics simulations.
- https://cdn.elifesciences.org/articles/101814/elife-101814-fig6-data3-v1.xlsx
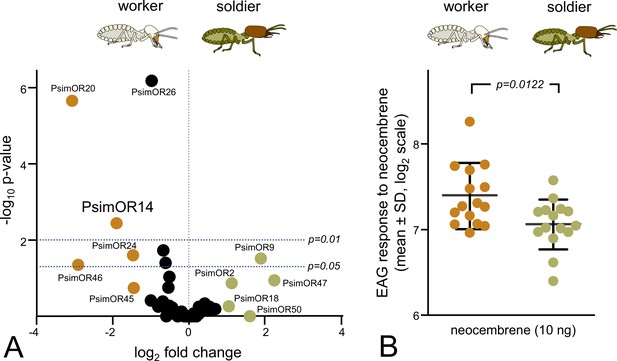
Caste comparison of PsimOR14 expression and EAG responses between P. simplex workers and soldiers.
(A) Volcano plot representing edgeR differential gene expression analysis of all 50 P. simplex odorant receptors (ORs) in RNAseq data from soldier and workers heads (including antennae) sequenced in three independent biological replicates per caste. Colored dots mark ORs reaching absolute value of log2 fold change ≥1, horizontal lines represent p-value thresholds of 0.05 and 0.01. Based on SRA archives accessible under SRX18952230–32 and SRX18952237–39. Numeric values of the edgeR and DESeq2 differential expression analysis are provided in Figure 7—source data 1. (B) EAG responses of whole antenna preparations of workers and soldiers to neocembrene at a dose of 10 ng (mean ± SD shown on log2 scale). Inter-caste differences were compared using a t-test on log2-transformed data. Raw data is shown in Figure 7—source data 2.
-
Figure 7—source data 1
Results of the edgeR and DESeq2 differential expression analyses.
- https://cdn.elifesciences.org/articles/101814/elife-101814-fig7-data1-v1.xlsx
-
Figure 7—source data 2
EAG measurements.
- https://cdn.elifesciences.org/articles/101814/elife-101814-fig7-data2-v1.xlsx
Tables
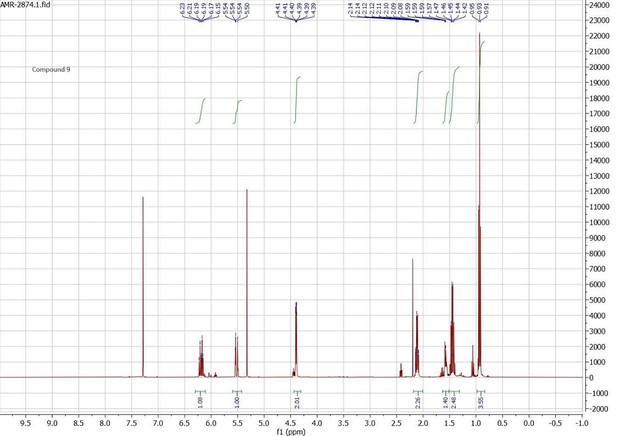
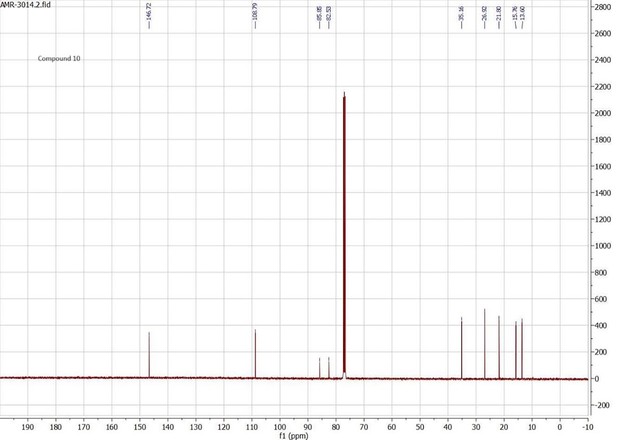
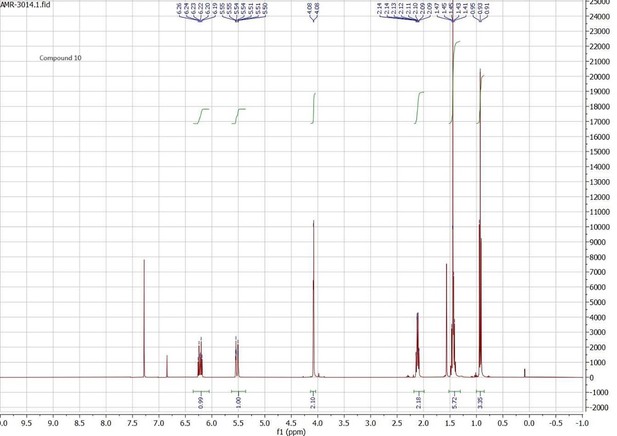
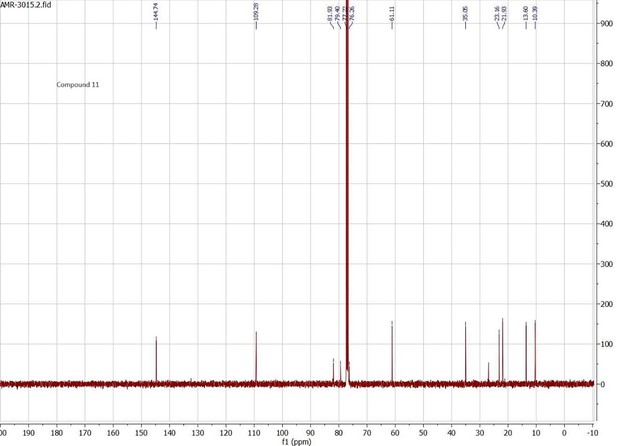
Docking scores and energy values inferred from the docking experiment and from MM/PBSA simulations for binding interactions of neocembrene, geranylgeraniol, and (+)-limonene with PsimOR14.
| Docking experiment | MM/PBSA E (kcal/mol) ± SD | ||||||
|---|---|---|---|---|---|---|---|
| Ligand | Docking score | VDWAALS | Electrostatic | ΔTOTAL | ΔVDWAALS | ΔEEL | ΔGSOLV |
| (kcal/mol) | |||||||
| Neocembrene | –8.658 | –19.777 | –0.223 | –28.72 ± 1.46 | –26.92 ± 1.24 | –0.29 ± 0.56 | –1.51 ± 0.10 |
| Geranylgeraniol | –8.331 | –18.786 | –11.137 | –36.98 ± 1.22 | –35.47 ± 0.96 | –0.77 ± 0.56 | –0.73 ± 0.27 |
| (+)-Limonene | –7.638 | –16.134 | –0.561 | –20.02 ± 2.63 | –20.57 ± 2.30 | –0.35 ± 0.47 | 0.89 ± 2.21 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Sequence-based reagent | PsimOR14_F | This paper | PCR primer – cloning | ATGATTCGATCAAAGAGAAAGG |
| Sequence-based reagent | PsimOR14_R | This paper | PCR primer – cloning | TTAGGAGTCGTGTAGATGAAT |
| Sequence-based reagent | PsimO31_F | This paper | PCR primer – cloning | ATGGAATACATAAAAAATGAAACATATTCTCA |
| Sequence-based reagent | PsimO31_R | This paper | PCR primer – cloning | TCAACCTACGACATGTGAGTTATT |
| Sequence-based reagent | PsimOR9_F | This paper | PCR primer – cloning | ATGGACAGCCTTTACGACCAATCTT |
| Sequence-based reagent | PsimOR9_R | This paper | PCR primer – cloning | TCATTCAGTGACTGAGGGATCCTT |
| Sequence-based reagent | PsimO30_F | This paper | PCR primer – cloning | ATGGAGCACAGGAAATACAAAGTGACAA |
| Sequence-based reagent | PsimO30_R | This paper | PCR primer – cloning | TTACGTTCCCTGATTTGTGTCGGTAT |
| Sequence-based reagent | PsimOrco_F | This paper | PCR primer – cDNA check | ATGTACAAGTTCAGGTTACACG |
| Sequence-based reagent | PsimOrco_R | This paper | PCR primer – cDNA check | CTAGTTGAGCTGTACCAACAC |
| Sequence-based reagent | GW1 | Thermo Fisher Scientific | PCR and Sanger sequencing primer | GTTGCAACAAATTGATGAGCAATGC |
| Sequence-based reagent | GW2 | Thermo Fisher Scientific | PCR and Sanger sequencing primer | GTTGCAACAAATTGATGAGCAATTA |
| Sequence-based reagent | UAS1 | Gonzalez et al., 2016 | Sanger sequencing primer | TAGCGAGCGCCGGAGTATAAATAG |
| Sequence-based reagent | UAS2 | Gonzalez et al., 2016 | Sanger sequencing primer | ACTGATTTCGACGGTTACCC |
| Sequence-based reagent | DmOr22a_F | This paper | PCR primer – genotyping | TCTCCAGCATCGCCGAGTGT |
| Sequence-based reagent | DmOr22a_R | This paper | PCR primer – genotyping | CGGCAGAGGTCCAGTCCGAT |
| Sequence-based reagent | PsimOR14_SW_F | This paper | PCR primer – genotyping | GAGAGCCAAGCAAACGAAAC |
| Sequence-based reagent | PsimOR14_SW_R | This paper | PCR primer – genotyping | TTTAGAAGGGAGCCACATCAC |
| Sequence-based reagent | PsimO31_SW_F | This paper | PCR primer – genotyping | GCTGGGTTAATCCCGATCAT |
| Sequence-based reagent | PsimO31_SW_R | This paper | PCR primer – genotyping | GCATGGCACCAAATAGTTCTTC |
| Sequence-based reagent | PsimOR9_SW_F | This paper | PCR primer – genotyping | TGGGCGAAACTGAGGATATG |
| Sequence-based reagent | PsimOR9_SW_R | This paper | PCR primer – genotyping | CGAGCCGACATAGAAGAAGAG |
| Sequence-based reagent | PsimO30_SW_F | This paper | PCR primer – genotyping | TGCCATCACCAGCAGATAAA |
| Sequence-based reagent | PsimO30_SW_R | This paper | PCR primer – genotyping | CACCGACTGACTCAGCATATT |
| Commercial assay or kit | PureLink RNA Mini | Invitrogen | Cat. #: 12183018A | |
| Commercial assay or kit | SuperScript IV Reverse Transcriptase | Invitrogen | Cat. #: 18090050 | |
| Commercial assay or kit | DreamTaq Green PCR Master Mix | Invitrogen | Cat. #: K1081 | |
| Commercial assay or kit | QIAquick Gel Extraction Kit | QIAGEN | Cat. #: 28706 | |
| Commercial assay or kit | pCR8/GW/TOPO TA Cloning Kit | Invitrogen | Cat. #: K250020 | |
| Strain, strain background (Escherichia coli) | OneShot TOP10 | Invitrogen | Cat. #: C404010 | Competent cells, Certificates of Analysis available at https://www.thermofisher.com/order/catalog/product/C404010 |
| Commercial assay or kit | QIAprep Spin Miniprep Columns | QIAGEN | Cat. #: 27115 | |
| Commercial assay or kit | Gateway LR Clonase Enzyme mix | Invitrogen | Cat. #: 11791019 | |
| Recombinant DNA reagent | pUASg.attb (plasmid) | Drosophila Genomics Resource Center, Bloomington, USA | DGRC Stock 1422; https://dgrc.bio.indiana.edu//stock/1422; RRID:DGRC_1422 | |
| Recombinant DNA reagent | pUASg.attB-PsimOR (plasmid) | This paper | ||
| Genetic reagent (D. melanogaster) | w; Or22abGAL4 | Thomas O. Auer (from Richard Benton Lab, University of Lausanne, Switzerland) | FLYB:FBal0018186 | Chahda et al., 2019 |
| Genetic reagent (D. melanogaster) | W1118 | Michal Žurovec (from Laboratory of Molecular Genetics, Institute of Entomology, Czechia) | ||
| Genetic reagent (D. melanogaster) | w-; Bl/Cyo; TM2/TM6B | MPI-Jena, Germany | ||
| Genetic reagent (D. melanogaster) | w-; +/+; UAS-OR(w+)/UAS-OR(w+) | This paper | On-demand commercial transgenesis by BestGene Inc, USA | |
| Sequence-based reagent (Cryptotermes secundus) | CsecOR and ORco sequences | Johny et al., 2023 | ||
| Sequence-based reagent (Zootermopsis nevadensis) | ZnevOR and ORco sequences | Johny et al., 2023 | ||
| Sequence-based reagent (Lepisma saccharina) | LsacOR sequences | Thoma et al., 2019 | ||
| Sequence-based reagent (Inquilinitermes inquilinus) | IinqOR and ORco sequences | Johny et al., 2023 | ||
| Sequence-based reagent (Neotermes cubanus) | NcubOR and ORco sequences | Johny et al., 2023 | ||
| Sequence-based reagent (Prorhinotermes simplex) | PsimOR and ORco sequences | Johny et al., 2023 |