Identifying genetic variations in emm89 Streptococcus pyogenes associated with severe invasive infections
Figures
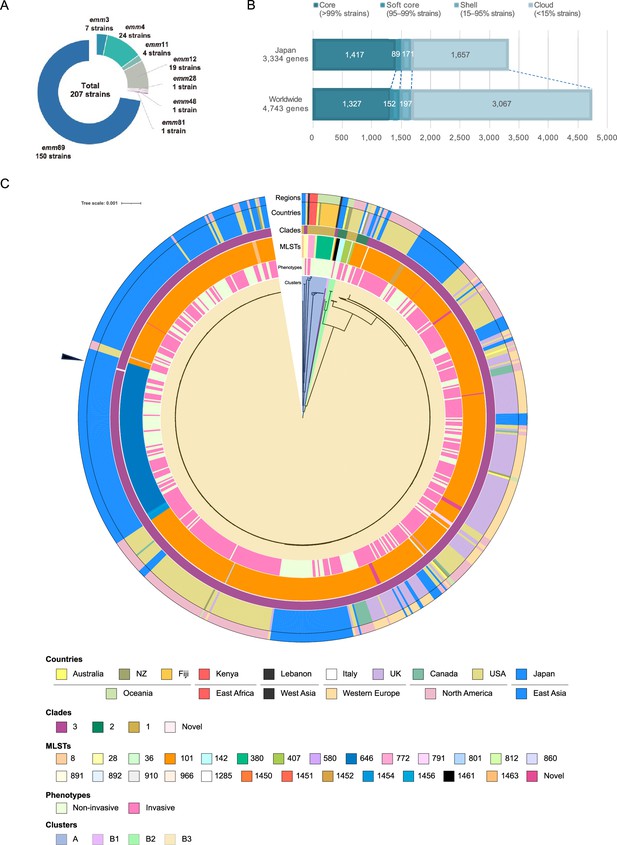
Pan-genomic and phylogenetic characterization of the collected emm89 S. pyogenes isolates.
(A) emm genotyping of the 207 clinical isolates collected in Japan. (B) Pan-genome analysis of the Japanese and global cohorts. All genes detected in each cohort were classified into four groups, according to prevalence: core, soft-core, shell, and cloud genes. (C) Phylogenetic tree for the global cohort, based on the sequences of the core genes. From the inside, the color bars show clusters, phenotypes, MLSTs, clades, and countries where strains were isolated, and global regions, in the order mentioned. The root of the tree was set as the mid-point. The scale located upper left indicates 0.001 times substitution of the bases on average. Arrowhead indicates strain belonging to the novel clade. MLST, multilocus sequence typing.
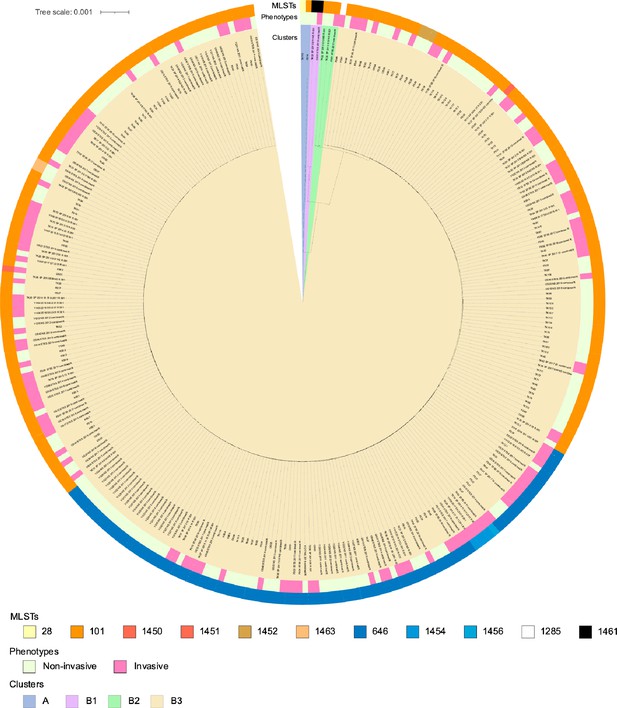
Phylogenetic tree for the Japanese cohort.
From the inside, color bars show clusters, phenotypes, and multilocus sequence typing (MLSTs), in the order mentioned. The root of the tree was set as the mid-point. The scale located in the upper left indicates 0.001 times substitution of the bases on average.
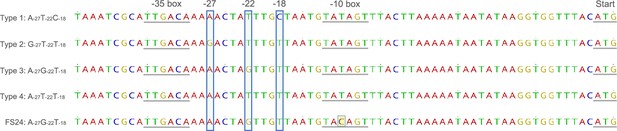
Variations in the nga promoter region.
Sequences of the nga promoter are classified into four major genotypes, according to three key residues (shown as −27,–22, and –18 in the figure) affecting promoter activity. Turner et al. further determined subtypes using mutations other than the key residues. FS24 strain has the same allele A−27G−22T−18 as the type 3 or clade 3 but has a single-nucleotide polymorphism (SNP) highlighted in yellow in the –10 box.
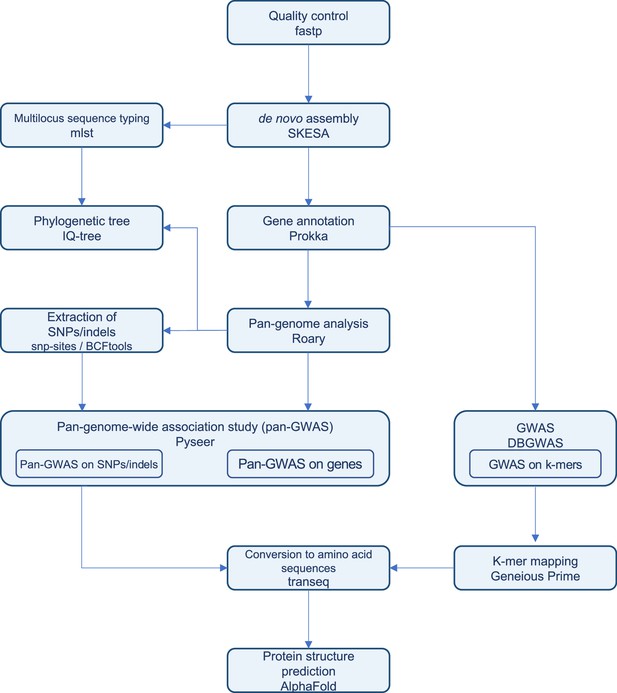
Pipeline for bacterial pan-genome analysis, pan-genome-wide association study (pan-GWAS), and GWAS constructed in this study.
We developed a pipeline for whole-genome analyses working on the National Institute of Genetics (NIG) Supercomputer and SQUID at the Cybermedia Center, Osaka University.
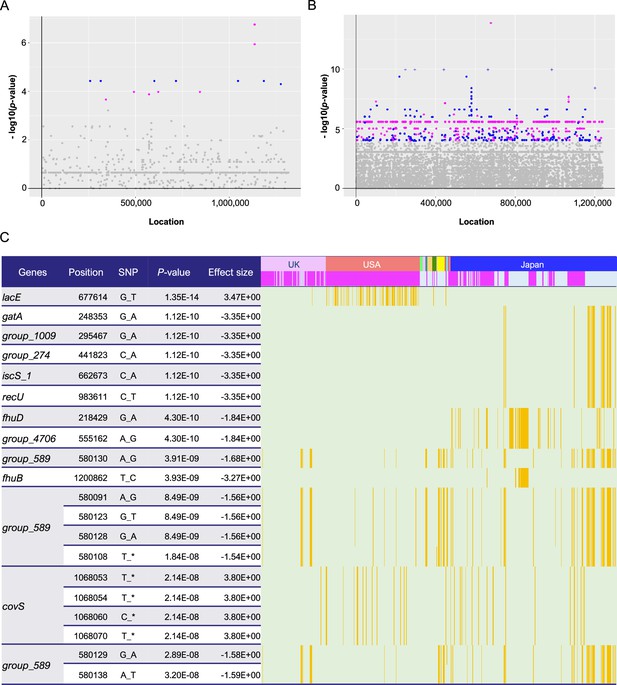
Pan-genome-wide association study on SNPs/indels.
Manhattan plots for the Japanese (A) and global (B) cohorts. The X-axis shows the location of each SNP/indel on the core gene alignment, while the Y-axis indicates the p-value. For each cohort, a permutation test was performed by iterating the calculations 1000 times with randomly permuted phenotypes, with the significance level set at the 5th percentile of the 1000 minimal p-values (p = 5.75 × 10–4 and p = 1.05 × 10–4 for the Japanese and global cohorts, respectively). Plots with lower p-values than genome-wide significant levels are colored magenta and blue, based on the direction of their effect size (positive and negative, respectively). (C) Distribution heatmap for the global cohort, with the strains possessing the significant SNPs/indels colored orange. Only the 20 SNPs with the lowest p-values are shown in this heatmap. Colored bars above indicate countries and phenotypes, and magenta bars represent invasive phenotypes. Using the Roary program, gene names starting with ‘Group_’ were automatically assigned. Position indicates the location of each SNP/indel on the core gene alignment. The full results are shown in Supplementary file 1, table S6. SNP, single-nucleotide polymorphism; indel, insertion/deletion.
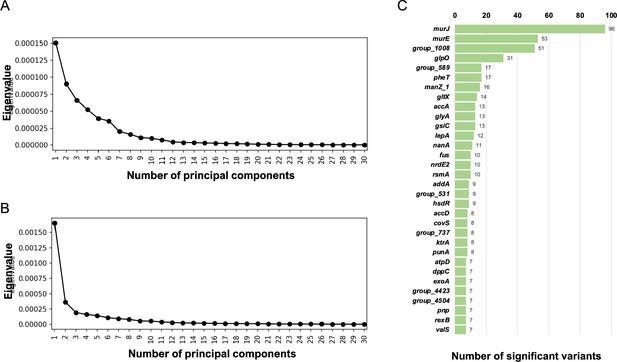
Pan-genome-wide association study (pan-GWAS) on single-nucleotide polymorphisms (SNPs)/indels.
Scree plot of eigenvalues generated by decomposition of the genetic distance matrix for the Japanese (A) and global (B) cohorts. To determine the number of principal components used as covariates for pan-GWAS, the plot in which the subsequent principal components seem to have relatively small eigenvalues was sought. Consequently, we used seven and three principal components as covariates for the Japanese and global cohorts, respectively. (C) Number of significant SNPs accumulated in single genes found in the pan-GWAS for the global cohort. Only the top 30 genes are shown in this graph.
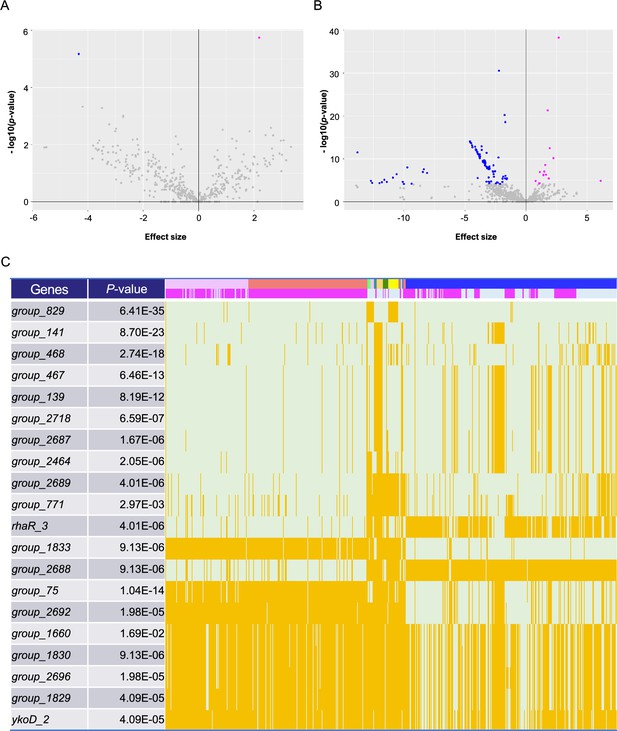
Pan-genome-wide association study on gene presence.
Volcano plots for the Japanese (A) and global (B) cohorts. The X-axis shows the effect size, while the Y-axis indicates the p-value. Plots with a lower p-value than the genome-wide significant levels (p = 1.09 × 10–4 and p = 7.72×10–5 for the Japanese and global cohorts, respectively) have been colored magenta and blue, based on the direction of their effect size (positive and negative, respectively). (C) Distribution heatmap for the global cohort, with the strains possessing the significant genes colored orange. Only the 20 genes with the lowest p-values are shown in this heatmap. Colored bars above indicate countries and phenotypes, and magenta bars represent invasive phenotypes. Using the Roary program, gene names starting with ‘Group_’ were automatically assigned. The full results are shown in Supplementary file 1, table S8.
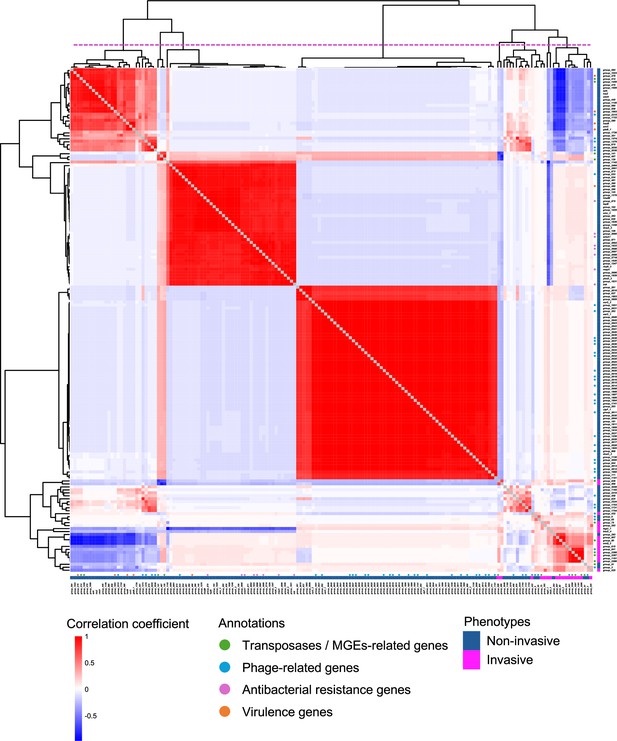
Pairwise correlation of significant clusters of orthologous genes (COGs).
Each cell indicates the correlation coefficient of each significant COG pair on its presence in the global cohort. The COGs are sorted based on hierarchical clustering. Annotations and phenotypes are represented by circles and cells located on the right and bottom of the heatmap. The dotted line on the tree divides the COGs into eight clusters.
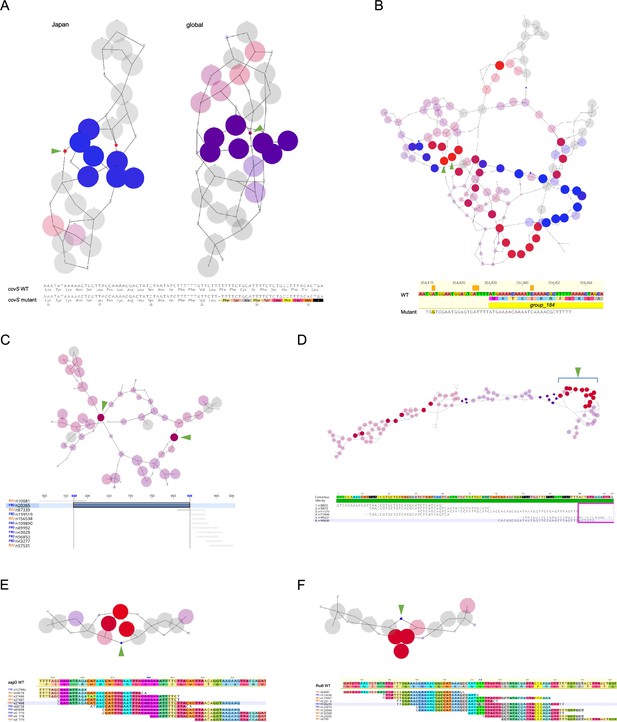
K-mers related to pathology.
Detailed results of the GWAS on k-mers for the Japanese cohort in two genomic regions: covS (A, top left) and group_184 (B); and the global cohort in five regions: covS (A, top right), an intergenic region (C), group_141–143 (D), sagG (E), and fhuB (F). (A–F, top) de Bruijn graphs generated using DBGWAS. The respective nodes in the graphs indicate k-mers, with the significant nodes indicated using green arrowheads. The size of each node corresponds to the allele frequency. (A–F, bottom) Alignments of k-mers or maps of k-mers on the genome sequence of MGAS27061 around regions including significant mutations. Each base is colored according to the interpreted amino acids. (A) The arrowheads indicate significant k-mers that cause a frameshift mutation, truncating CovS protein to 35 amino acids. (B) The significant k-mers indicate that the presence of a sequence mapped on the first 26 bp of group_184 and its upstream 20 bp is related to the pathology. (C) Two significant k-mers indicate the same intergenic region of 270 bp. (D) The significant k-mers are indicated using arrowheads.
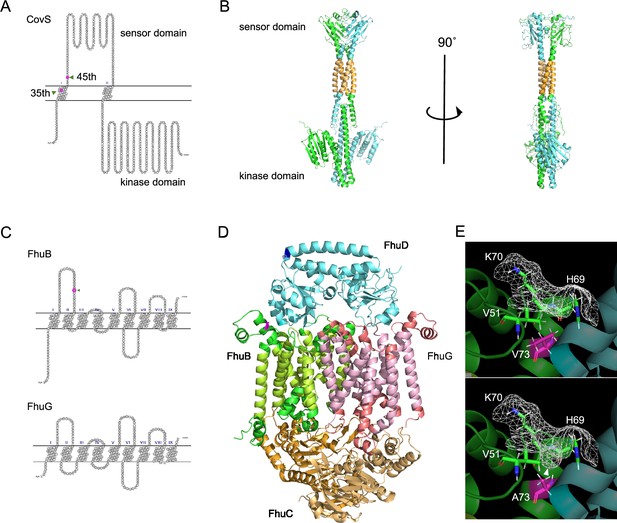
Predicted protein structure models.
(A) Snake-like plot of transmembrane regions of CovS estimated using SOSUI. Frameshift mutations, detected by the single-nucleotide polymorphisms (SNPs)/indel- and k-mer-based GWASes, cause truncation of CovS at the indicated 35th and 45th residues, respectively. (B) Structural model of the CovS homodimer (ipTM + pTM = 0.614). The putative transmembrane regions are colored orange. The upper part is the sensor domain, while the lower is the C-terminal kinase domain involved in the phosphorylation of the transcriptional regulator CovR. (C) Snake-like plot depicting the transmembrane regions of FhuB and FhuG. The 73rd residue of FhuB is indicated by an arrowhead and in magenta. (D) Structural model of the FhuBDCCG complex (ipTM + pTM = 0.791). The putative transmembrane regions of FhuB and FhuG are colored green-yellow and peach, respectively. The upper part of the model is located in the extracellular region, while the lower part is in the cytoplasm. (E) The 73rd valine in FhuB, shown in magenta, was substituted with alanine. The molecular surface is illustrated with a wireframe, and that of the predicted indentation is shown with an arrowhead. ipTM + pTM: Weighted combination of interface-predicted TM and predicted TM scores. ipTM is used to measure structural accuracy in the protein–protein interface, while pTM is a metric for overall topological accuracy.
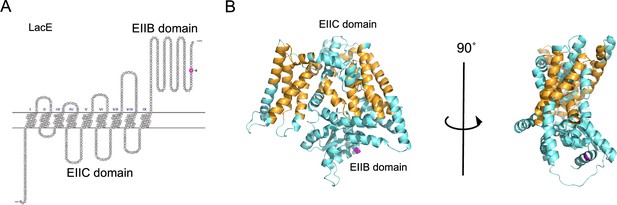
Predicted protein structure model of LacE.
(A) Snake-like plot of transmembrane regions of LacE estimated using SOSUI. The 554th residue has been indicated with an arrowhead and in magenta. (B) Structure model of LacE generated with AlphaFold (pLDDT = 0.902). The putative transmembrane regions are colored in orange. LacE has two domains: the transmembrane domain EIIC and intracellular domain EIIB. The 554th glycine substituted with valine is colored in magenta. pLDDT: The best predicted local difference distance test indicating local structural accuracy.
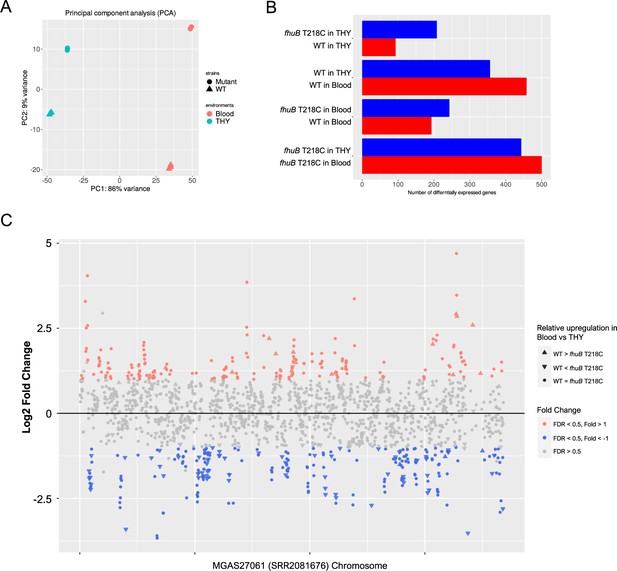
Transcriptome analysis of the fhuB T218C mutant strain in THY and human blood.
(A) Principal component analysis plot of RNA-seq data. (B) Differentially expressed genes in four comparisons. (C) Plot of gene expression in the WT strain versus fhuB T218C mutant in human blood. Significantly up- and downregulated genes in the WT are colored red and blue, respectively. The shapes of the plots indicate relative transcriptional changes between THY and blood. Genes depicted with upward triangles are either significantly upregulated in the WT or downregulated in the mutant strain, in blood versus THY. The downward triangle plots indicate genes that are either downregulated in the WT or upregulated in the mutant strain, in blood versus THY. WT, wild-type; THY, Todd Hewitt broth supplemented with 0.2% yeast extract.
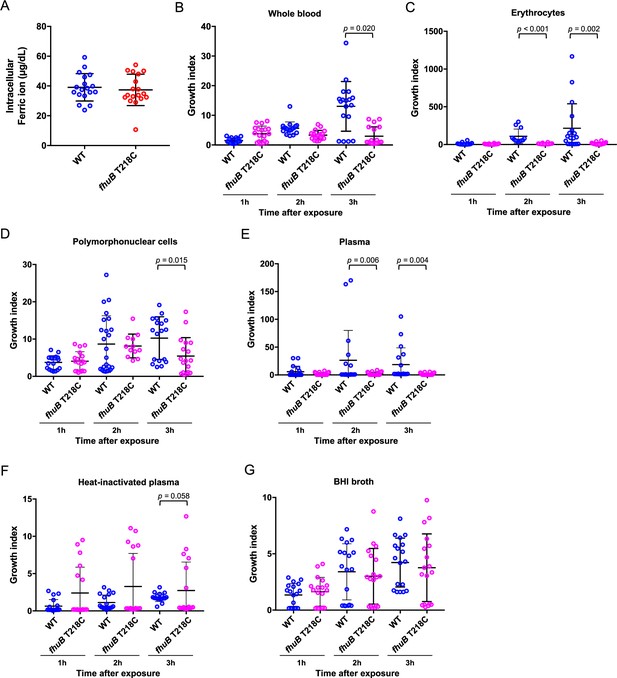
Effects of the fhuB T218C mutant strain on ferric ion uptake and bacterial survival in human blood.
(A) Intracellular ferric ion assay. The wild-type (WT) and fhuB T218C mutant strains were incubated in healthy human blood for 3 hr, following which the intracellular ferric ion concentrations were measured. (B) Bactericidal assay in human blood. The WT and fhuB T218C mutant were mixed with healthy human blood to measure the ratio of bacterial counts at 1, 2, and 3 hr to those at 0 h after infection. Bactericidal assay in human erythrocyte-rich medium (C), polymorphonuclear cell-rich medium (D), plasma (E), and plasma inactivated by heating at 56°C for 30 min (F). (G) Bacterial growth in brain heart infusion broth. The data were pooled from three independent experiments, each performed in sextuplicate. Thick bars and error bars indicate means and quartiles, respectively. Statistical significance was determined using the Mann–Whitney test with Benjamini–Hochberg’s correction.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Streptococcus pyogenes) | TK02′ | This paper | Clinical isolate from severe invasive infection; wild-type reference for mutagenesis (see Materials and methods) | |
| Strain, strain background (Streptococcus pyogenes) | TK02′-fhuB T218C | This paper | Isogenic mutant strain carrying fhuB T218C point mutation (see Materials and methods) | |
| Strain, strain background (Escherichia coli) | DH5α | Takara Bio | Host strain for plasmid construction | |
| Chemical compound, drug | Carbenicillin | Nacalai Tesque | Used for selection in E. coli | |
| Chemical compound, drug | Spectinomycin | Fujifilm Wako Pure Chemical Corporation | Used for selection in E. coli and S. pyogenes | |
| Chemical compound, drug | Mutanolysin | Sigma-Aldrich | Used for bacterial lysis | |
| Chemical compound, drug | Lysozyme | Fujifilm Wako Pure Chemical Co | Used for bacterial lysis | |
| Chemical compound, drug | Achromopeptidase | Fujifilm Wako Pure Chemical Co | Used for bacterial lysis | |
| Chemical compound, drug | RNase A | Promega | Used for RNA degradation during DNA extraction | |
| Commercial assay or kit | Maxwell RSC instrument | Promega | Automated DNA extraction | |
| Commercial assay or kit | Nextera XT DNA Kit | Illumina | Library preparation for sequencing | |
| Commercial assay or kit | QuantiChrom Iron Assay Kit | BioAssay Systems | Used for intracellular ferric ion measurement | |
| Software, algorithm | Fastp v0.20.1 | Chen et al., 2018 | RRID:SCR_016962 | Used for quality filtering of sequencing reads |
| Software, algorithm | SKESA v2.4.0 | Souvorov et al., 2018 | Used for de novo genome assembly | |
| Software, algorithm | MLST v2.19.0 | Used for multi-locus sequence typing | ||
| Software, algorithm | Prokka v1.14.5 | Seemann, 2014 | RRID:SCR_014732 | Used for genome annotation |
| Software, algorithm | Roary v3.12.0 | Page et al., 2015 | RRID:SCR_018172 | Used for pan-genome analysis |
| Software, algorithm | IQ-tree v1.6.12 | Nguyen et al., 2015 | RRID:SCR_017254 | Used for phylogenetic tree construction |
| Software, algorithm | pyseer v1.3.4 | Lees et al., 2018 | Used for pan-GWAS | |
| Software, algorithm | DBGWAS v0.5.3 | Jaillard et al., 2018 | Used for k-mer-based GWAS | |
| Software, algorithm | AlphaFold v2.2.2 | Jumper et al., 2021 | RRID:SCR_025454 | Used for protein structure prediction |
| Software, algorithm | Geneious Prime v2022.0.1 | Biomatters | RRID:SCR_010519 | Used for sequence visualization and mapping |
| Software, algorithm | STAR v2.7.0a | RRID:SCR_004463 | Used for RNA-seq read mapping | |
| Software, algorithm | featureCounts v1.5.2 | Liao et al., 2014 | RRID:SCR_012919 | Used for RNA-seq read counting |
| Software, algorithm | iDEP v0.96 | Ge et al., 2018 | Used for transcriptomic data analysis | |
| Software, algorithm | Prism v7.0c | GraphPad | RRID:SCR_002798 | Used for statistical analysis |
| Commercial assay or kit | Quick-RNA Miniprep Kit | Zymo Research | Used for RNA extraction from bacterial samples |
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/101938/elife-101938-mdarchecklist1-v1.docx
-
Supplementary file 1
Tables S1–S16.
- https://cdn.elifesciences.org/articles/101938/elife-101938-supp1-v1.xlsx
-
Source data 1
Sequencing data for the fhuB T218C mutant strain.
- https://cdn.elifesciences.org/articles/101938/elife-101938-data1-v1.zip





