Crystal structure and catalytic mechanism of PL35 family glycosaminoglycan lyases with an ultrabroad substrate spectrum
Figures
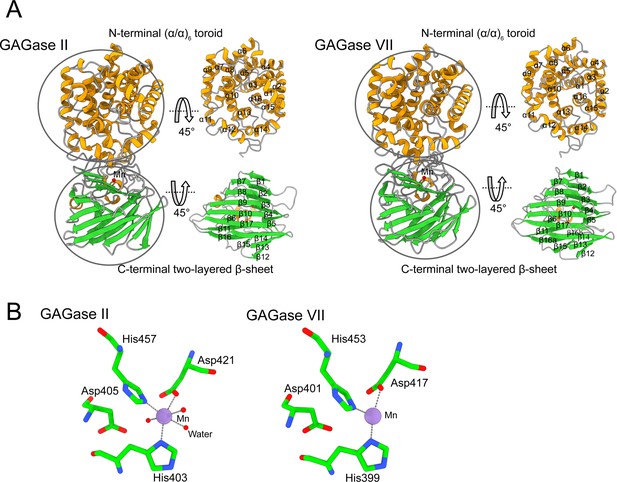
Structural description of glycosaminoglycan lyases (GAGase) II and GAGase VII.
(A) Overall structures of GAGase II (left) and GAGase VII (right) are shown in carton. The α-helix, β-strand, and random coil are colored with yellow, green, and gray, respectively. The N-terminal (α/α)6 toroid domain and C-terminal two-layered β-sheet domain were circled and the secondary structure elements are marked nearby. (B) Mn binding site of GAGase II and GAGase VII. The purple sphere presents Mn2+. Mn2+ binding site of GAGase II (left) and GAGase VII (right) is shown in stick. A cut-off distance of 3.0 Å was carried out to choose neighboring residues.
Structural modeling and alignment of other glycosaminoglycan lyases (GAGases).
The structures of the other GAGases were predicted by RoseTTAfold. All of them show good superposition in their predicted structures.
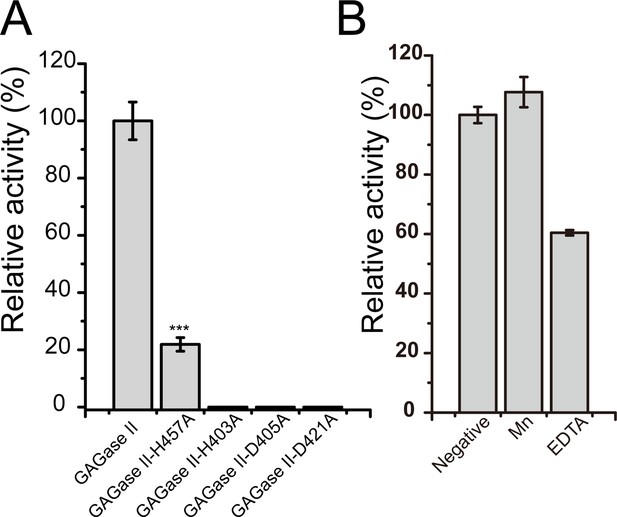
Activity of glycosaminoglycan lyases (GAGase II) and its variants.
(A) Activity of site-directed mutants of His and Asp nearby the Mn2+. His and Asp residues nearby the Mn2+ were individually mutated to Ala and the relative activity of each variant was measured as described in ‘Materials and methods.’ All of the residual activities were evaluated and shown as the relative intensity compared with that of wild-type GAGase II. (B) The effects of Mn2+ and chelating reagent of GAGase II were determined using hyaluronan (HA) (1 mg/ml) as substrate. Error bars represent averages of triplicates (n=3) ± S.D. by Student’s t test; ***p<0.001.
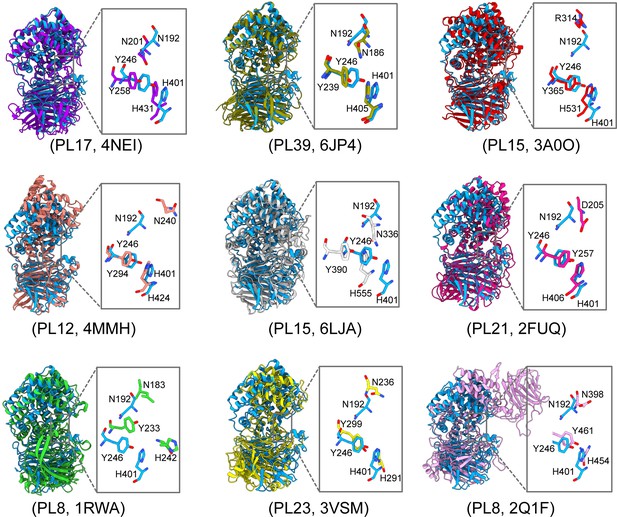
Multiple structural alignments of glycosaminoglycan lyases (GAGase) II and its structurally similar proteins.
GAGase II (8KHV, blue) was aligned with structurally identified glycosaminoglycans (GAGs) and alginate lyases, including PL17 family alginate lyase (4NEI, purple), PL39 family alginate lyase (6JP4, olive), PL15 family alginate lyase (3A0O, red), PL12 family heparinase III (4MMH, salmon), PL15 family exoHep (6LJA, gray), PL21 family heparinase II (2FUQ, magenta), PL8 family chondroitin sulfate AC lyase II (1RWA, green), PL23 family chondroitinase (3VSM, yellow), and PL8 family chondroitin sulfate ABC lyase II (2Q1F, pink). The detailed views of the crucial active site residues are shown in stick mode. The root-mean-square deviation (RMSD) between these structures and GAGase II were calculated as 1.34, 1.34, 1.30, 1.24, 1.15, 1.35, 1.24, 1.14, and 1.41 Å based on 171, 148, 141, 111, 99, 87, 60, 22, and 21 pruned atoms, respectively.
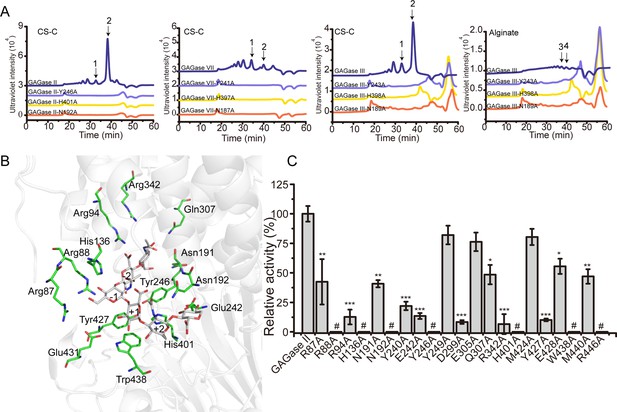
Catalytic center and substrate binding sites of glycosaminoglycan lyases (GAGases).
(A) The crucial catalytic site-directed mutagenesis of GAGase II, GAGase III, and GAGase VII. The conserved crucial residues of GAGase II, GAGase III, and GAGase VII were individually mutated to Ala. CS-C and alginate were used as substrates for the activity evaluation of GAGase II, GAGase III, GAGase VII, and its variants. The activity of each variant was detected using gel filtration HPLC on a Superdex Peptide column as described in ‘Materials and methods;’ the elution of each fraction is indicated as follows: 1, CS-C tetrasaccharide; 2, CS-C disaccharide; 3, alginate trisaccharide; 4, alginate disaccharide. (B) Molecular docking of GAGase II with a hyaluronan (HA) hexasaccharide. The molecule docking was carried out with GAGase II and a HA hexasaccharide (PDB code: 1HYA) to predict the substrate binding sites. The binding site residues (green) and hexasaccharide ligand (gray) are showed as sticks. (C) The putative substrate-binding site residues surrounding the docking substrate were individually mutated to Ala. HA was treated with each variant at 40 °C for 12 hr and the relative activity of each variant was shown as the relative intensity compared with that of wild-type GAGase II. Error bars represent averages of triplicates (n=3) ± S.D. by Student’s t test; *p<0.01, **p<0.001, ***p<0.0001, #: the activities of the variants were too low to be accurately detected and almost completely inactivated.
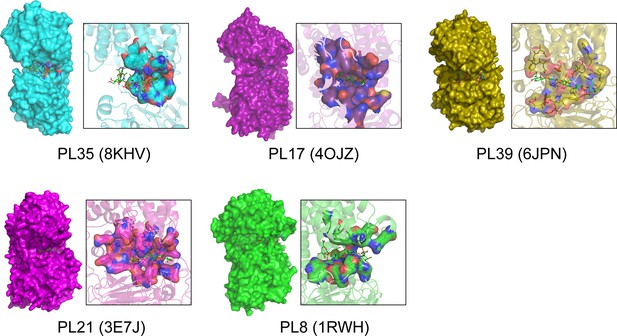
Surface representations and substrate binding tunnels of glycosaminoglycan lyases (GAGase) II and its structurally similar glycosaminoglycan (GAG)/alginate lyase.
The surface representation of GAGase II (8KHV, blue) from PL35 family, alginate lyase from PL17 family (4OJZ, purple, ligand: β-D-mannopyranuronic acid-(1-4)-α-D-mannopyranuronic acid-(1-4)-α-L-gulopyranuronic acid), alginate lyase from PL39 family (6JPN, olive, ligand: β-D-mannopyranuronic acid-(1-4)-β-D-mannopyranuronic acid-(1-4)-β-D-mannopyranuronic acid-(1-4)- β-D-mannopyranuronic acid-(1-4)-β-D-mannopyranuronic acid), heparinase II from PL21 family (3E7J, magenta, ligand: 4-deoxy-α-L-threo-hex-4-enopyranuronic acid-(1-4)–2-acetamido-2-deoxy-β-D-glucopyranose-(1-4)-α-D-glucopyranuronic acid-(1-4)–2-acetamido-2-deoxy-β-D-glucopyranose), and chondroitinase AC II from PL8 family (1RWH, green, ligand: 6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-3)–2-acetamido-2-deoxy-4-O-sulfo-β-D-galactopyranose-(1-4)–2,6-anhydro-3-deoxy-L-xylo-hexonic acid-(1-3)–2-acetamido-2-deoxy-4-O-sulfo-β-D-galactopyranose). The dimensions of their substrate-binding cavities are measured as follows:14.24 Å (PL35, 8KVI, Asp233-OD2 to Tyr246-OH), 24.25 Å (PL17, 4OJZ, Lys94-NZ to Gln138-NE2), 22.97 Å (PL39, 6JPN, Arg183-NH2 to Tyr343-OH), 25.55 Å (PL21, 3E7J, Glu136-ZN to Lys197-ZN) and 16.97 Å (PL8, 1RWH, Arg134-NH2 to Arg174-NH1).
SDS-PAGE of glycosaminoglycan lyases (GAGases) and their variants.
The expression and purification of the indicated GAGases and their various mutants, including the putative substrate binding site variants of GAGase II (A), the key triplet residues variants of GAGase II/III/VII (B), and other related variants (C), were assessed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) followed by staining with Coomassie Brilliant Blue R-250.
-
Figure 4—figure supplement 2—source data 1
The original files of the full raw uncropped, unedited gels.
- https://cdn.elifesciences.org/articles/102422/elife-102422-fig4-figsupp2-data1-v1.zip
-
Figure 4—figure supplement 2—source data 2
Figures with the uncropped gels or blots with the relevant bands clearly labeled.
- https://cdn.elifesciences.org/articles/102422/elife-102422-fig4-figsupp2-data2-v1.zip
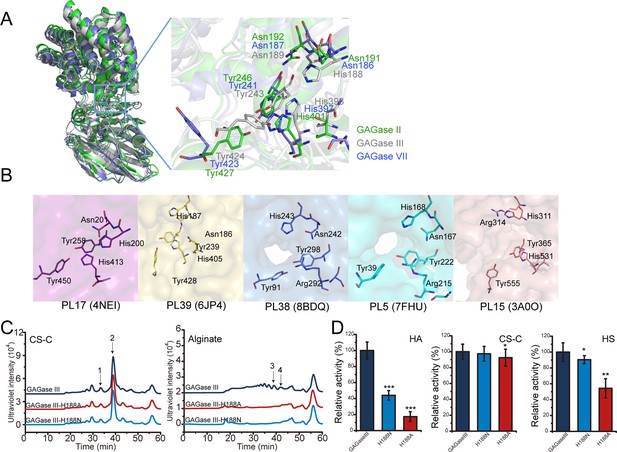
Analysis of a key residue for the alginate-degrading activity of glycosaminoglycan lyases (GAGase) III.
(A), Multiple structural alignment of GAGase II, III, and VII. GAGase II (8KHV, green) and GAGase VII (8KHW, skyblue) were aligned with GAGase III (model, gray). The detailed views of crucial catalytic residues are shown in stick mode; (B), Conserved catalytic residues of alginate lyases from PL17 (4NEI, purple), PL39 (6JP4, yellow), PL38 (8BDQ, slate), PL5 (7FHU, cyan) and PL15 (3A0O, pink) family. Residues are shown in stick; (C), Activity assay of GAGase III-H188N and GAGase III-H188A toward CS-C and alginate. The crucial site His188 was mutated to alanine and asparagine, respectively. The activity of each variant against CS-C and alginate was assessed using gel filtration HPLC on a Superdex Peptide column, as described under ‘Materials and methods;’ the elution of each fraction is indicated as follows: 1, CS-C tetrasaccharide; 2, CS-C disaccharide; 3, alginate trisaccharide; 4, alginate disaccharide; (D), Relative activity of GAGase III and its variants. Three types of GAGs (HA, CS-C, and HS) were individually treated with GAGase III and its variants (GAGase III-H188N and GAGase III-H188A) at 40 °C for 1 hr; relative activities of enzymes were determined by detecting the absorbance at 232 nm. Data are shown as the percentage of the activity relative to the wild-type GAGase III. Error bars represent averages of triplicates (n=3) ± S.D. by Student’s t test; *p<0.5; **p<0.01; ***p<0.001.
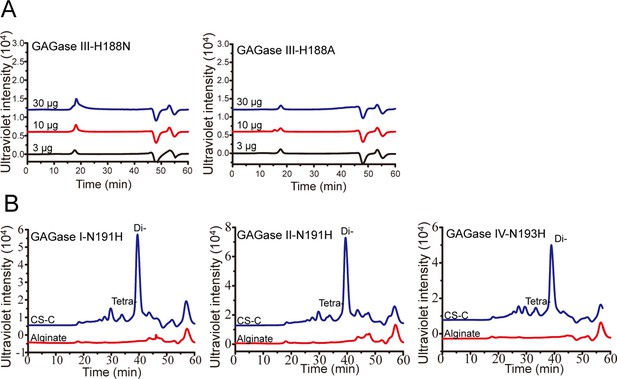
Activity assay of glycosaminoglycan lyases (GAGases) variants against CS-C and alginate.
(A), Activity assay of different concentrations of GAGase III-H188N and GAGase III-H188A against alginate; (B), Activity assay of GAGase I-N191H, GAGase II-N191H, and GAGase IV-N193H, where the crucial asparagine residues in GAGase I, II, and IV were mutated to histidine, respectively. The activities of the corresponding mutants against CS-C or alginate were evaluated using gel filtration HPLC on a Superdex Peptide column as described under ‘Materials and methods;’ Di- is indicated as CS disaccharide; Tetra- is indicated as chondroitin sulfate (CS) tetrasaccharide.
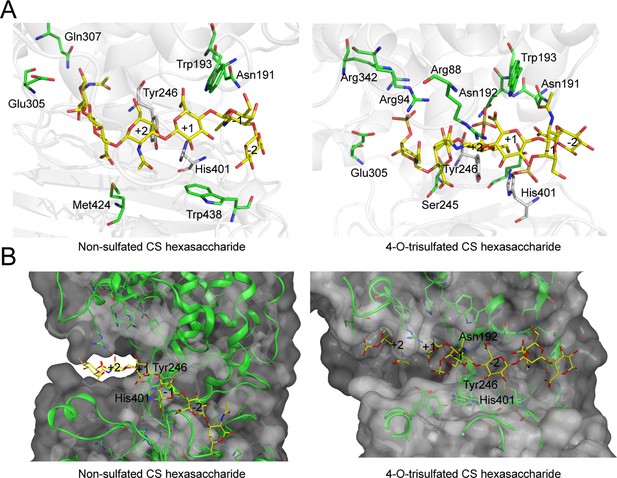
Molecular docking of glycosaminoglycan lyases (GAGase) II with hexasaccharide ligands.
Molecular docking of GAGase II with chondroitin sulfate (CS) ligands. The molecule docking was carried out with GAGase II and CS ligands, including nonsulfated (PDB code: 2KQO) (left) and 4-O-trisulfated (PDB code: 1C4S) (right) CS hexasaccharide, to investigate the substrate selectivity, using AutoDock Vina (A) and Molecular Operating Environment (MOE) (B). The catalytic triplet residues (green) and CS ligand (yellow) are showed as sticks.
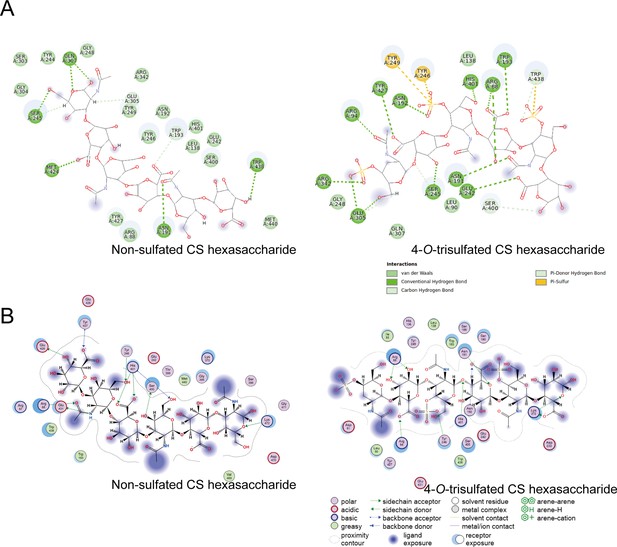
wo-dimensional interaction plot of glycosaminoglycan lyases (GAGase) II with molecular docking substrates.
(A), GAGase II form hydrogen bonds, carbon-hydrogen bonds, Pi hydrogen bonds, Pi-Sulfur, and van der Waals interactions with nonsulfated (PDB code: 2KQO) (left) and 4-O-trisulfated (PDB code: 1C4S) (right) chondroitin sulfate (CS) hexasaccharides. Interactions were analyzed by discovery studio software. (B), Interaction between GAGase II and nonsulfated CS hexasaccharide (PDB code: 2KQO) (left) or 4-O-trisulfated CS hexasaccharide (PDB code: 1C4S) (right) was analyzed using Molecular Operating Environment (MOE).
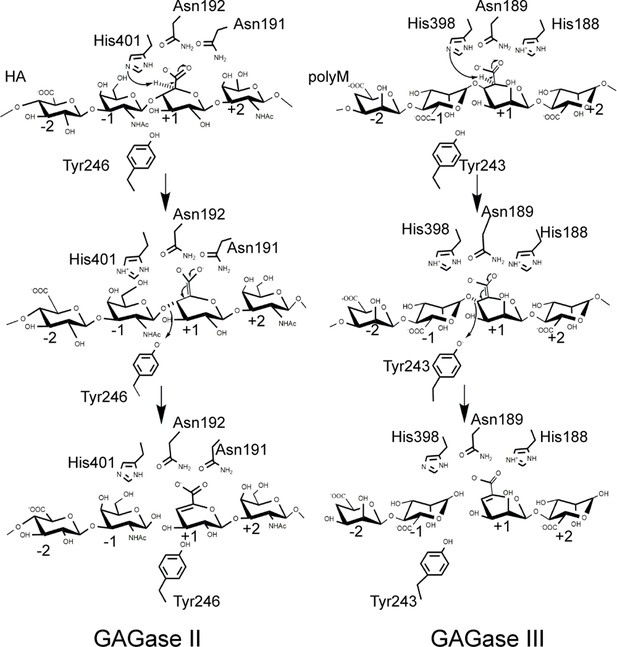
Proposed catalytic mechanism of glycosaminoglycan lyases (GAGase) II and GAGase III.
Take the hyaluronan (HA) degradation by GAGase II and the polyM degradation by GAGase III for example. Briefly, the substrate is firstly binding to the negatively charged residues near the catalytic sites, the carboxylate group is neutralized by Asn192, Asn191 in GAGase II and His188, Asn189 in GAGase III; His398 in GAGase II and His401 in GAGase III are proposed to work as general base to abstract a proton from the C5 position of GlcUA at +1 position, and Tyr246 in GAGase II and Tyr243 in GAGase III work as general acid to donate the leaving group at –1 position a proton. The arrows indicate the direction of electron transfer, and the C5-C6 double bond on the middle panel indicate the enolate anion intermediate created by proton abstraction at C5 position.
Tables
Data collection and refinement statistics.
| GAGase II | GAGase VII | SeMet-GAGase II | |
|---|---|---|---|
| PDB entry code | 8KHV | 8KHW | - |
| Data collection | |||
| Wavelength (Å) | 0.98 | 0.98 | 0.98 |
| Resolution range | 44.01–1.90 (1.94–1.90) | 19.87–2.40 (2.49–2.40) | 40.91–2.00 (2.07–2.00) |
| Space group | P21 | P43212 | P21 |
| Unit cell | a=45.92 Å b=82.10 Å c=79.89 Å α=90º β=106.59º γ=90º | a=98.41 Å b=98.41 Å c=134.75 Å α=90º β=90º γ=90º | a=46.39 Å b=81.82 Å c=79.66 Å α=90º β=106.88º γ=90º |
| Unique reflections | 86421 (4092) | 49305 (2737) | 38212 (3738) |
| Completeness (%) | 98.55 (92.04) | 99.68 (99.85) | 99.15 (98.47) |
| Rmeas | 0.10 (0.23) | 0.10 (0.74) | 0.08 (0.24) |
| Rp.i.m | 0.04 (0.10) | 0.02 (0.16) | 0.03 (0.093) |
| Mean I/σ(I) | 12.9 (6.1) | 20.6 (4.3) | 17.9 (8.2) |
| CC1/2 | 0.99 (0.96) | 0.99 (0.97) | 0.99 (0.97) |
| Refinement statistics | |||
| Rwork | 0.16 (0.17) | 0.2079 (0.26) | - |
| Rfree | 0.1919 (0.23) | 0.2722 (0.36) | - |
| RMSD bond length (Aº) | 0.006 | 0.008 | - |
| RMSD bond angle (º) | 0.79 | 0.99 | - |
| Ramachandran plot (%) | |||
| Favored | 97.75 | 94.31 | - |
| Allowed | 2.25 | 5.00 | - |
| Outliers | 0.00 | 0.69 | - |
| Rotamer outliers (%) | 0.00 | 0.40 | - |
| Clashscore | 2.78 | 6.89 | - |
| Average B-factor (Å2) | 13.51 | 54.14 | - |
| Macromolecules | 11.66 | 54.20 | - |
| Ligands | 21.71 | 45.54 | - |
| Solvent | 25.75 | 47.24 | - |
-
Statistics for the highest-resolution shell are shown in parentheses.
Structural similarity of glycosaminoglycan lyases (GAGase) II/GAGase VII with glycosaminoglycan (GAG)/alginate lyases analyzed using DALI.
| Chain | Z-score | RMSD (Å) | lali | nres | Description | PL family | |
|---|---|---|---|---|---|---|---|
| GAGase II | 3a0o-A | 29.4 | 3.2 | 539 | 764 | Alginate lyase | 15 |
| 6jp4-A | 28.9 | 3.0 | 523 | 770 | Alginate lyase | 39 | |
| 4nei-A | 27.3 | 3.1 | 523 | 705 | Alginate lyase | 17 | |
| 4mmh-A | 28.6 | 4.4 | 481 | 637 | Heparinase III | 12 | |
| 6lja-A | 25.7 | 3.9 | 540 | 841 | Exo-type heparinase | 15 | |
| 2fuq-A | 25.1 | 4.9 | 528 | 743 | Heparinase II | 21 | |
| 1rwa-A | 15.2 | 5.4 | 470 | 754 | Chondroitin AC lyase | 8 | |
| 3vsm-a | 14.3 | 5.3 | 369 | 633 | Chondroitinase | 23 | |
| 2q1f-a | 11.1 | 5.3 | 434 | 991 | Chondroitin ABC lyase | 8 | |
| GAGase VII | 3a0o-A | 29.2 | 3.6 | 545 | 764 | Alginate lyase | 15 |
| 6jp4-A | 28.0 | 3.3 | 530 | 770 | Alginate lyase | 39 | |
| 4nei-A | 27.2 | 3.2 | 529 | 705 | Alginate lyase | 17 | |
| 4mmh-A | 26.4 | 4.4 | 492 | 637 | Heparinase III | 12 | |
| 2fuq-A | 23.1 | 5.4 | 531 | 747 | Heparinase II | 21 | |
| 1rwa-A | 14.3 | 4.5 | 473 | 754 | Chondroitin AC lyase | 8 | |
| 3vsm | 12.9 | 4.3 | 365 | 633 | Chondroitinase | 23 | |
| 2q1f-A | 10.5 | 4.6 | 407 | 991 | Chondroitin AC lyase | 8 |
Multiple structural alignments of (α/α)n toroid domain or antiparallel β-sheet domain of glycosaminoglycan lyases (GAGase) II and identified glycosaminoglycan (GAG)/alginate lyases.
| PL family | Domain | Aligned atoms | RMSD | Substrate | ||
|---|---|---|---|---|---|---|
| (α/α)n toroid domain | 8KHV | PL35 | (α/α)6 toroid (Met34-Trp350) | 317 | 0.00 | HA, CS, and HS |
| 3A0O | PL15 | (α/α)6 toroid (Gly127-Leu452) | 101 | 1.04 | Alginate | |
| 4NEI | PL17 | (α/α)6 toroid (His30-Glu364) | 102 | 1.20 | Alginate | |
| 6JP4 | PL39 | (α/α)6 toroid (Ala1-Tyr355) | 113 | 1.30 | Alginate | |
| 2FUQ | PL21 | (α/α)6 toroid (Thr27-Leu368) | 67 | 1.42 | Hep, HS | |
| 1RWA | PL8 | (α/α)6 toroid (Pro4-Val367) | 60 | 1.24 | HA, CS | |
| 3VSM | PL23 | (α/α)5 toroid (Asn72- Asn330) | 22 | 1.14 | CS | |
| 7FHU | PL5 | (α/α)6 barrel (Cys23-Pro329) | 27 | 1.105 | Alginate | |
| 8BDQ | PL38 | (α/α)7 barrel (Ala24-Lys402) | 58 | 1.177 | Alginate | |
| 4MMH | PL12 | (α/α)5 toroid (Ile29-Thr379) | 24 | 1.169 | Hep, HS | |
| antiparallel β-sheet domain | 8KHV | PL35 | anti-parallel β-sheet | 261 | 0.00 | HA, CS, and HS |
| 3A0O | PL15 | anti-parallel β-sheet | 109 | 1.11 | Alginate | |
| 4NEI | PL17 | anti-parallel β-sheet | 85 | 0.97 | Alginate | |
| 6JP4 | PL39 | anti-parallel β-sheet | 67 | 1.06 | Alginate | |
| 2FUQ | PL21 | anti-parallel β-sheet | 103 | 1.17 | Hep, HS | |
| 4MMH | PL12 | anti-parallel β-sheet | 97 | 1.22 | Hep, HS |
Catalytic residues comparison of identified glycosaminoglycans (GAGs) and alginate lyases shared similar structure with glycosaminoglycan lyases (GAGase) II and GAGase VII.
| PDB code | PL family | Substrates | Sequence identity(Query cover/Per. Ident) * | Catalytic residues | ||
|---|---|---|---|---|---|---|
| General base/acid | Neutralizer | |||||
| GAGase II | 8KHV | PL35 | HA, CS, and HS | 100%/100% | Tyr246, His401 | Asn192 |
| GAGase VII | 8KHW | PL35 | HA, CS, and HS | 93%/44.67% | Tyr241, His397 | Asn187 |
| GAGase III | - | PL35 | HA, CS, HS, and alginate (M-specific) | 98%/64.08% | Tyr243, His398, | Asn189, His188 |
| Chondroitin sulfate AC lyase II | 1RWA | PL8 | HA and CS | N.D.† | His233, Tyr242 | Asn183 |
| Chondroitin sulfate ABC lyase II | 2Q1F | PL8 | HA, CS, and DS | N.D. | His345, Tyr461, His454 | Asp398 |
| Heparinase III | 4MMH | PL12 | HS | 8%/30.91% | Tyr294, His424 | Asn240 |
| Alginate lyase | 3A0O | PL15 | Alginate (M/G specific) | 43%/23.93% | His311, Tyr365, His531 | Arg314 |
| Exo-Heparinase | 6LJA | PL15 | Hep and HS | N.D. | His337, Tyr390, His555 | Asn336 |
| Alginate lyase | 4NEI | PL17 | Alginate (M-specific) | 15%/25.51% | Tyr258, His413 | Asn201, His200 |
| Heparinase II | 2FUQ | PL21 | Hep and HS | 29%/23.13% | His202, Tyr257, His406 | Glu205 |
| Chondroitin lyase | 3VSM | PL23 | CS | N.D. | Tyr299, His291 | Asn236 |
| Alginate lyase | 6JP4 | PL39 | Alginate (M/G specific) | 31%/26.53% | Tyr239, His405 | Asn186, His187 |
-
*
Sequence identity means the sequence similarity compared with GAGase II.
-
†
N.D. means no significant identity is detected.
Additional files
-
Supplementary file 1
Supplementary data for crystal structure and catalytic mechanism of PL35 family glycosaminoglycan lyases with an ultrabroad substrate spectrum.
(a) Sequence information of the identified glycosaminoglycan lyases (GAGases). (b) Inductively coupled plasma-mass spectrometry (ICP-MS) analysis of GAGase II and GAGase VII. (c) Strains and primers used in this study.
- https://cdn.elifesciences.org/articles/102422/elife-102422-supp1-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/102422/elife-102422-mdarchecklist1-v1.pdf





