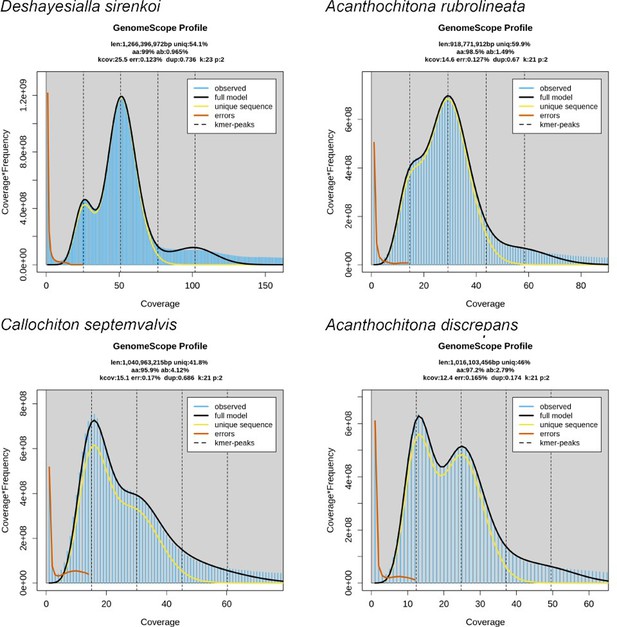
Still waters run deep in large-scale genome rearrangements of morphologically conservative Polyplacophora
Figures
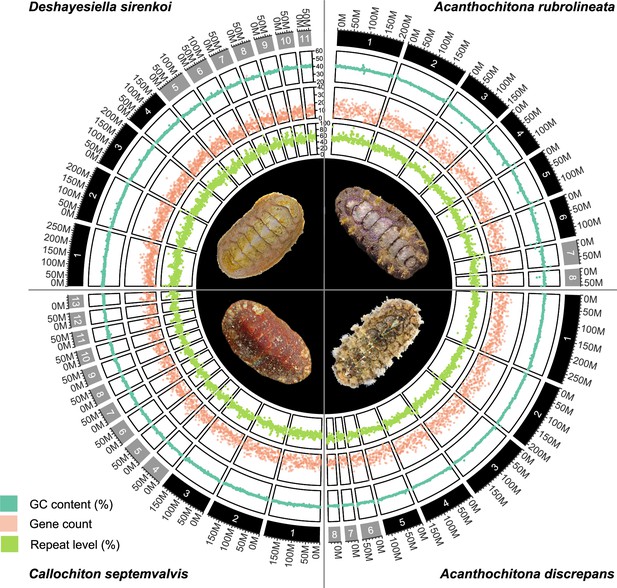
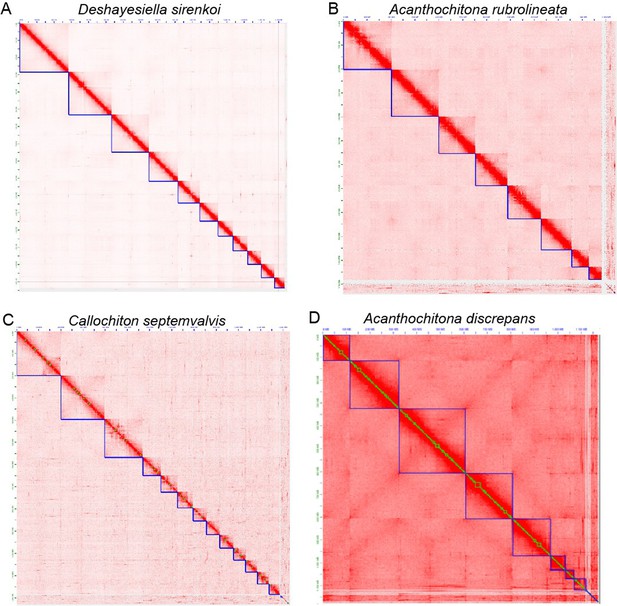
CIRCOS plots for four new chitons genome assemblies, clockwise from top left: one species in the order Lepidopleurida Deshayesiella sirenkoi, and three species in the clade Chitonida sensu lato: Acanthochiton rubrolineata, A.discrepans, and Callochiton septemvalvis.
Each quarter circle shows the pseudochromosome content for each species, in order of size, with concentric rings indicating GC content, gene count, percent repeat content, and a photograph of the respective species.
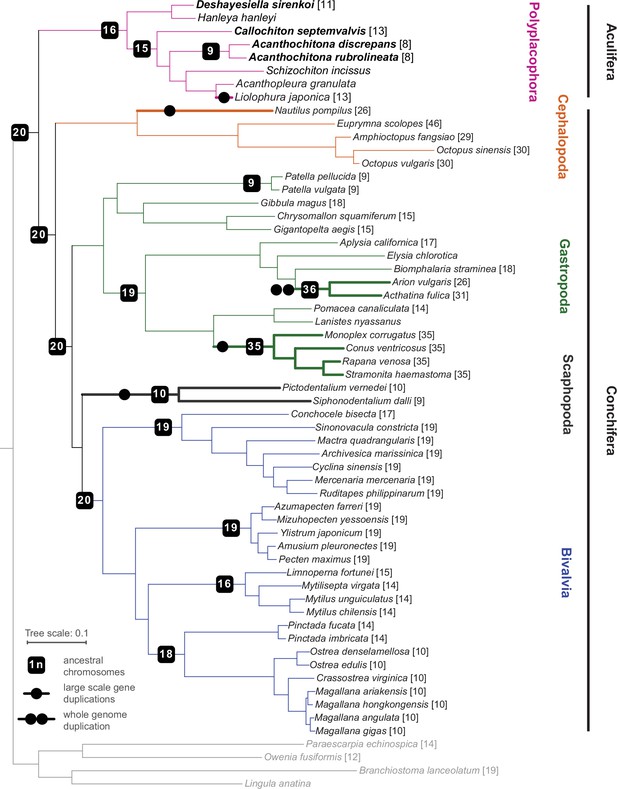
Phylogeny of Mollusca, with new genomes noted in bold type, and the chromosome number in square brackets for each species where known.
Lineages with known whole (double circle) or partial genome (single circle) duplication are in thicker lines for emphasis; boxes on branches show the reconstructed ancestral 1 n chromosome number for the respective clade.
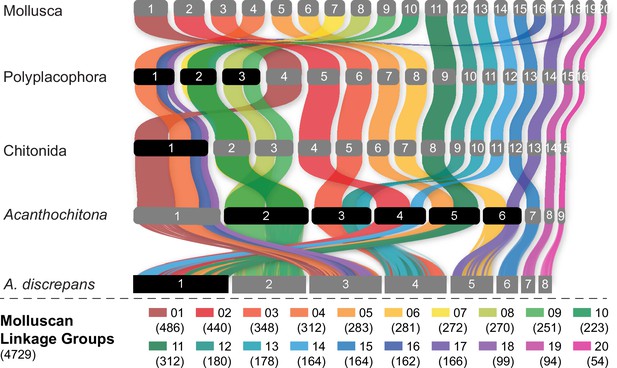
Evolution of the ancestral molluscan linkage groups (MLGs) within Polyplacophora using the lineage leading to Acanthochitona discrepans as an example.
MLGs are distinguished by colours, at the top of the diagram and in the key at bottom showing the number of orthologs. Each row is the reconstructed karyotype of the ancestor of living Polyplacophora, the order Chitonida sensu lato, and the genus Acanthochitona. Reconstructed chromosomes on each row are numbered in order of size from largest (left) to smallest (right); chromosome fusions are highlighted with chromosome numbers in black boxes. This presentation highlights the extent of shifts especially in comparison to the molluscan or polyplacophoran ancestor.
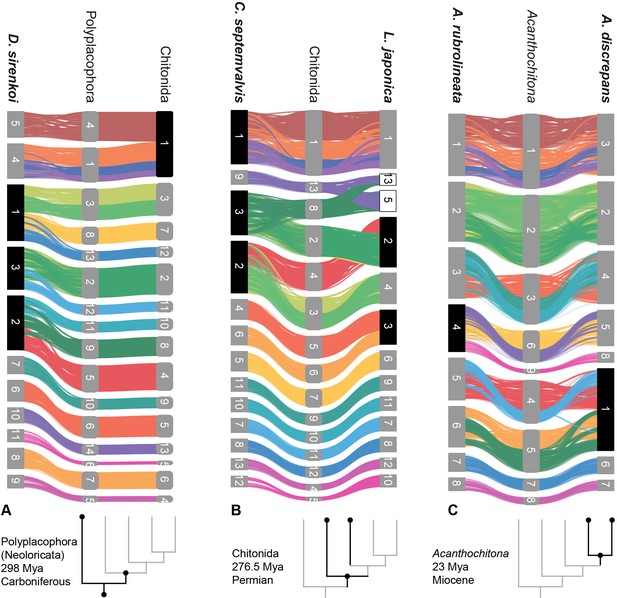
Syntenic rearrangements of MLGs within the evolution of Polyplacophora.
Each part shows the reconstructed karyotypes of an ancestor (middle) and two descendent lineages, with a schematic cladogram for orientation. From left to right, the divergence of A ancestor of Polyplacophora, leading to the lepidopleuran species Deshayesiella sirenkoi (left) and ancestor of Chitonida (right), B ancestor of Chitonida s.l. leading to the callochitonid Callochitona septemvalvis (left) and chitonid Liolophura japonica (right) and C ancestor of the genus Acanthochitona leading to the two congeneric species A. rubrolineata (left) and A. discreapans (right). Colours and presentation are as in Figure 3, and chromosome numbers indicate the sequence in terms of size from largest (1) to smallest. Here, the chromosomes are not in order of size but reordered such that each transition from the nearest ancestor is visible in more detail. Chromosome fusions are highlighted with chromosome numbers in black boxes, and duplications in white boxes.

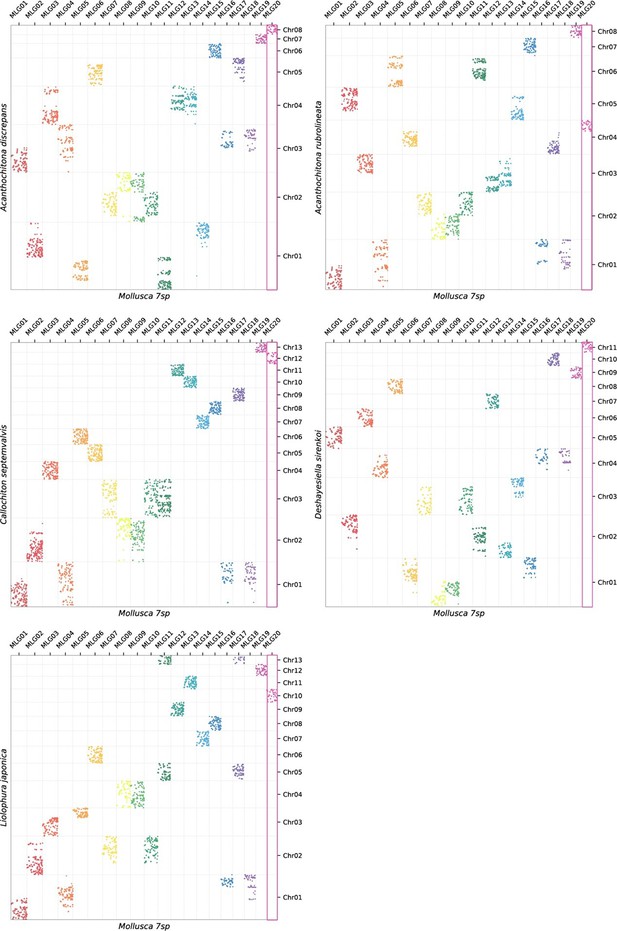
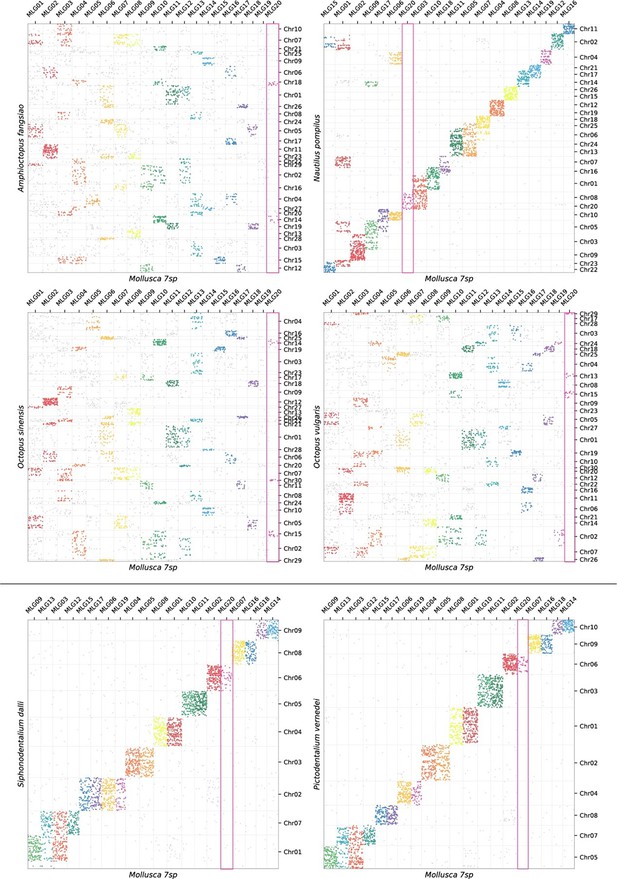
Oxford plots comparing gene occupancy of species from four different classes of molluscs (vertical) to the ancestral molluscan linkage groups (MLG, horizontal): (A) the bivalve Archivesica marisinica retains a plesiomorphic karyotype reflecting the 20 MLGs, (B) the chiton Liolophura japonica has large scale gene duplication in MLG11 and MLG17 on separate pseudochromosomes, (C) the scaphopod Siphonodetalium dalli has large scale gene duplication of MLG13 and MLG03 on separate pseudochromosomes, (D) the gastropod Rapana venosa demonstrates a nearly whole genome duplication with MLG20 not duplicated.
Colours follow the presentation in Figure 3.
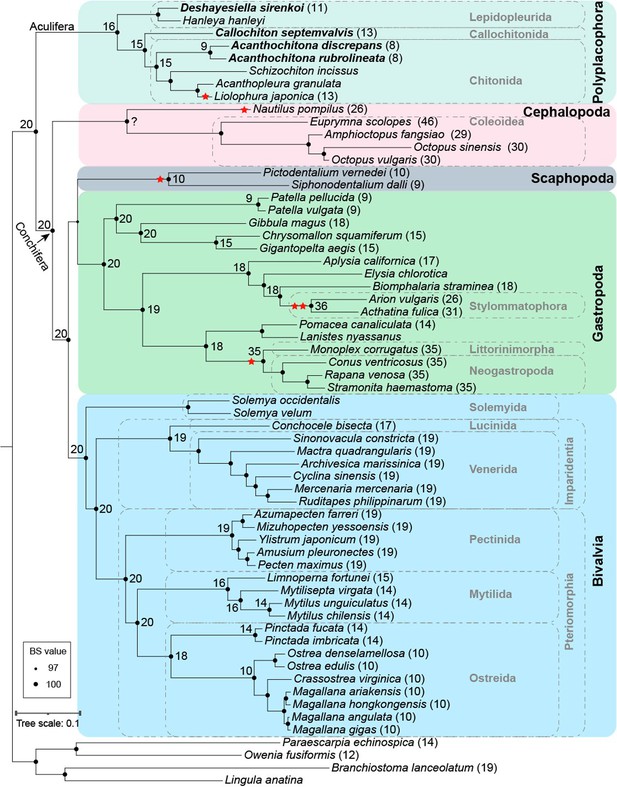
Phylogenomic relationships among five classes in Mollusca (with two Solemya from RNA-seq).
A single star in the tree indicates partial duplications and two stars indicate true whole genome duplication. The numbers in parentheses after taxon names indicate the number of pseudo-chromosomes where known, and the numbers on branches indicate the predicted (1 n) number of ancestral chromosomes. The newly sequenced chiton species are indicated in bold text. In contrast to the tree presented in the main text, this tree resolved Scaphapoda +Gastropoda. IQTREE2: -m MFP -B 1000 (60 genomes and 2 transcriptomes).
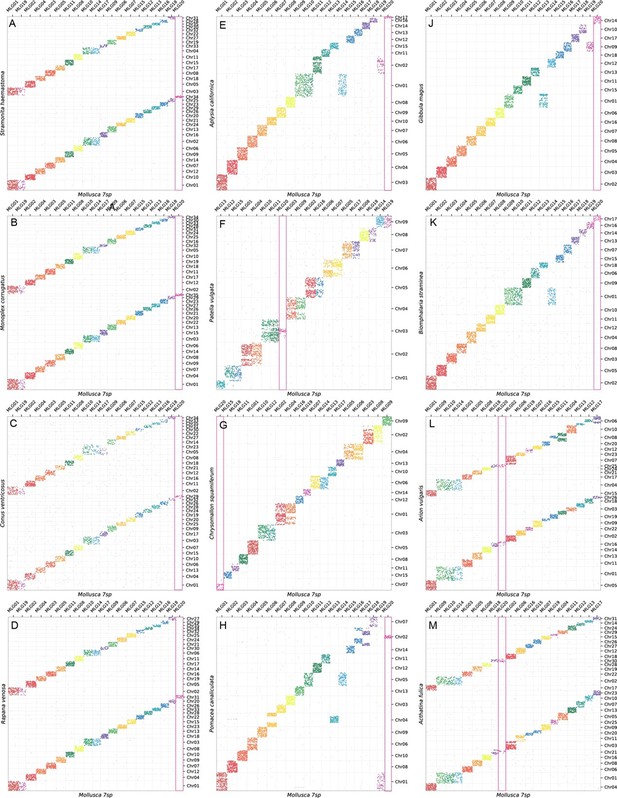
Oxford plots of 12 gastropods against MLGs, with the identification of the true sense of whole genome duplications (WGD).
MLG20 is highlighted with a red box to distinguish its dynamics under WGD.
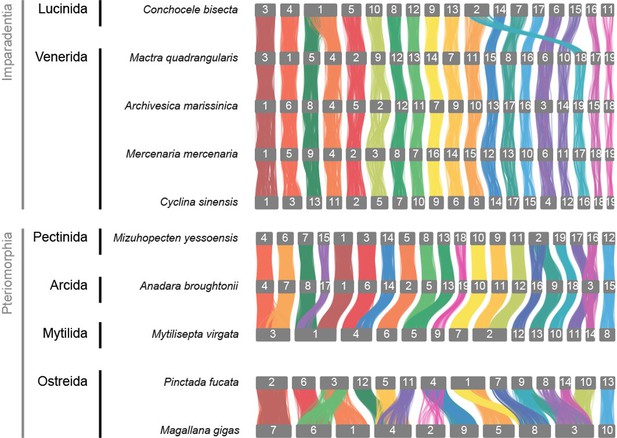
Syntenic graph in Bivalvia, highly conserved synteny in Imparadentia and large rearrangements within Peteriomorphia.
Two clades in Bivalvia have 19 pseudo-chromosomes from a single fusion, but they are independent occurrences: in Pectinida, the fusion is MLG12 and MLG16, whereas in Venerida, the fusion is MLG16 and MLG18.
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/102542/elife-102542-mdarchecklist1-v1.docx
-
Supplementary file 1
Supplementary tables.
Table S1. Summary statistics from genome sequencing of four new chiton genomes, including HiFi and Hi-C. Table S2. Genome assemblies and gene-model prediction for four new chiton genomes. Table S3. Comparison of summary data among available chitons genomes; the four new chiton genomes are indicated in bold text. N.B. analyses also incorporate the other chromosome-level genome for Liolophura japonica, which was published prior to the present study. Table S4. Nonsyntenic rate between species (summarised by clade in Table S5) following the method described in the Supporting Information appendix. Table S5. Summary information for translocation rate of 25 species of molluscs, organised by taxonomic class.
- https://cdn.elifesciences.org/articles/102542/elife-102542-supp1-v1.xlsx