Bacteriophage infection drives loss of β-lactam resistance in methicillin-resistant Staphylococcus aureus
Figures
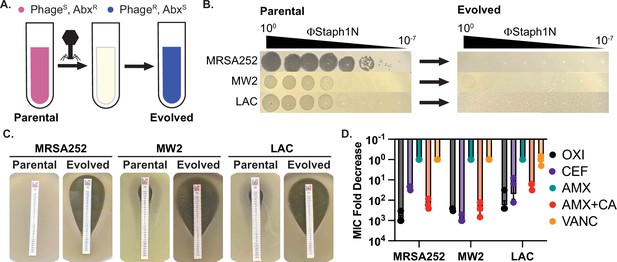
Infection by bacteriophage ΦStaph1N drives the loss of β-lactam resistance in MRSA.
(A) Schematic of the experimental setup. Drug-resistant (AbxR), phage-sensitive (PhageS) bacterial cultures are infected with phage. The population of infected cells is passaged and allowed to recover. The surviving cell population is resistant to phage infection (PhageR) but has evolved sensitivity to antibiotics (AbxS). (B) ΦStaph1N infects MRSA strains MRSA252, MW2, and LAC (left panel). Following infection with ΦStaph1N, evolved cultures of the three MRSA strains are resistant to ΦStaph1N (right panel). (C) ΦStaph1N-treated, evolved MRSA strains show significant loss of resistance against oxacillin, compared to the parental strains. Loss of resistance is indicated by the area of bacterial clearance surrounding the antibiotic resistance strip. (D) ΦStaph1N treatment causes loss of resistance against different β-lactams. Plotted are the fold reductions of minimal inhibitory concentration (MIC) between treated and mock-treated cells. OXI = oxacillin; CEF = cefazolin; AMX = amoxicillin; AMX + CA = amoxicillin and clavulanic acid; VANC = vancomycin. Error bars represent the Standard Error of the Mean (SEM) of three independent replicates.
-
Figure 1—source data 1
Uncropped plate images for Figure 1B and C.
- https://cdn.elifesciences.org/articles/102743/elife-102743-fig1-data1-v1.zip
-
Figure 1—source data 2
Source data for the bar graphs in Figure 1D.
- https://cdn.elifesciences.org/articles/102743/elife-102743-fig1-data2-v1.xlsx
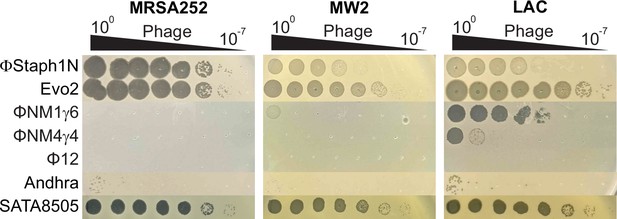
Phage sensitivity of MRSA strains.
Efficiencies of plaquing of phages on MRSA252, MW2, and LAC. Phages were 10-fold serially diluted and spotted onto top agar overlays of each strain.
-
Figure 1—figure supplement 1—source data 1
Uncropped plate images of Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/102743/elife-102743-fig1-figsupp1-data1-v1.zip
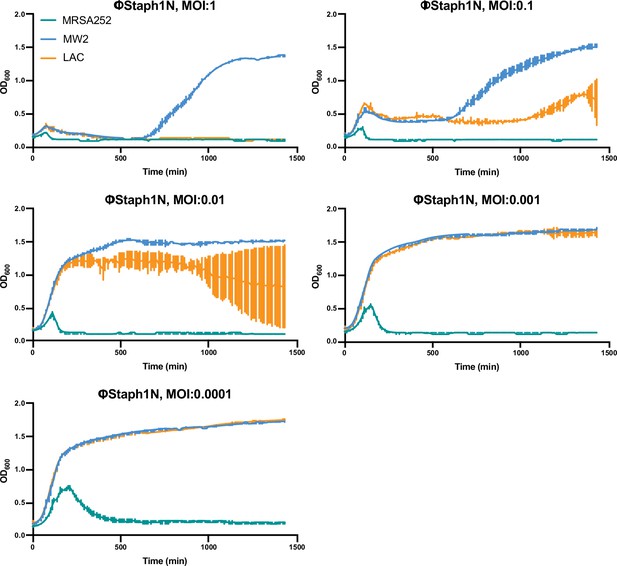
Growth curves of MRSA strains under varying levels of ΦStaph1N infection.
MRSA252, MW2, and LAC cultures were infected with ΦStaph1N at the indicated multiplicity of infection (MOI). The optical density (OD600) of the cultures was monitored on an automated plate reader. Each condition was tested in three independent replicates and error bars represent the Standard Deviation (SD).
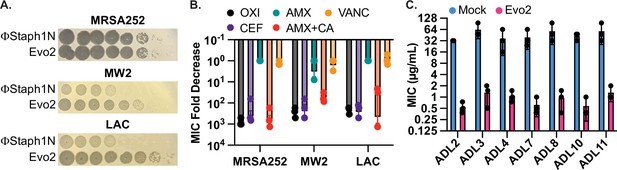
Evo2 is a variant of ΦStaph1N with higher activity against MRSA.
(A) Evo2 shows comparable infectivity towards MRSA252 but improved infectivity towards MW2 and LAC, relative to ΦStaph1N. The same plaquing data is also shown in Figure 2—figure supplement 1. (B) Similar to ΦStaph1N, Evo2 infection reduces β-lactam resistance in MRSA. (C) Evo2 infection reduces the MIC against oxacillin in clinical isolates of USA300 (ADLs). All error bars represent the Standard Error of the Mean (SEM) of three independent replicates.
-
Figure 2—source data 1
Uncropped plate images for Figure 2A.
- https://cdn.elifesciences.org/articles/102743/elife-102743-fig2-data1-v1.zip
-
Figure 2—source data 2
Source data for the bar graphs in Figure 2B and C.
- https://cdn.elifesciences.org/articles/102743/elife-102743-fig2-data2-v1.xlsx
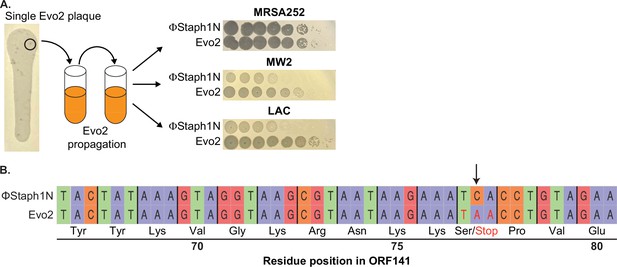
Isolation and sequencing analysis of Evo2.
(A) Individual Evo2 plaques appeared in larger ΦStaph1N plaques on LAC. Individual plaques were isolated and propagated in liquid culture. Evo2 shows improved plaquing on MW2 and LAC. Plaquing data in the right panels are the same as in Figure 2A. (B) Evo2 is a mutant form of ΦStaph1N with a nonsense mutation in ORF141. The A to C mutation (marked by the arrow) in Evo2 converts Serine 77 of ORF141 into a stop codon.
-
Figure 2—figure supplement 1—source data 1
Uncropped plate images for Figure 2—figure supplement 1A.
- https://cdn.elifesciences.org/articles/102743/elife-102743-fig2-figsupp1-data1-v1.zip
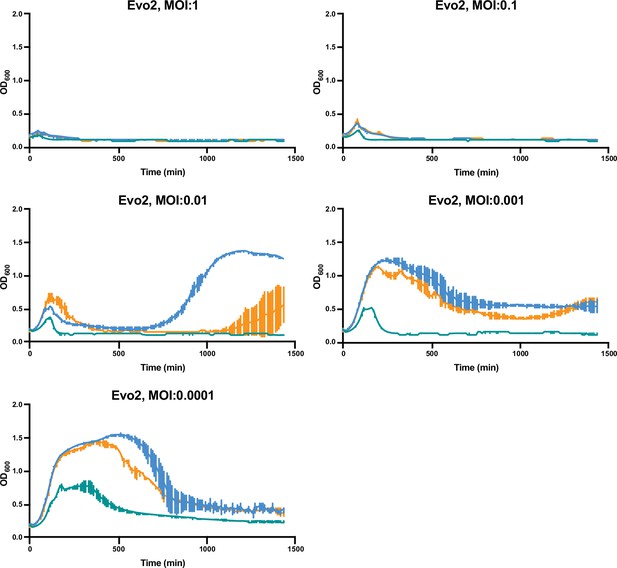
Growth curves of MRSA strains under varying levels of Evo2 infection.
MRSA252, MW2, and LAC cultures were infected with Evo2 at the indicated multiplicity of infection (MOI). The optical density (OD600) of the cultures was monitored on an automated plate reader. Each condition was tested in three independent replicates and error bars represent the Standard Deviation (SD).
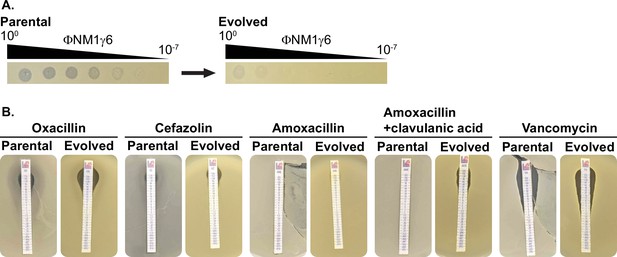
Phage ΦΝΜ1γ6 infection LAC does not drive the loss of β-lactam resistance.
(A) LAC treated with ΦΝΜ1γ6 evolves resistance against ΦΝΜ1γ6, evidenced by the reduction of plaquing from the parental to the evolved populations. (B) Evolved and parental LAC populations show comparable MICs against different β-lactams and vancomycin.
-
Figure 2—figure supplement 3—source data 1
Uncropped plate images for Figure 2—figure supplement 3A and B.
- https://cdn.elifesciences.org/articles/102743/elife-102743-fig2-figsupp3-data1-v1.zip
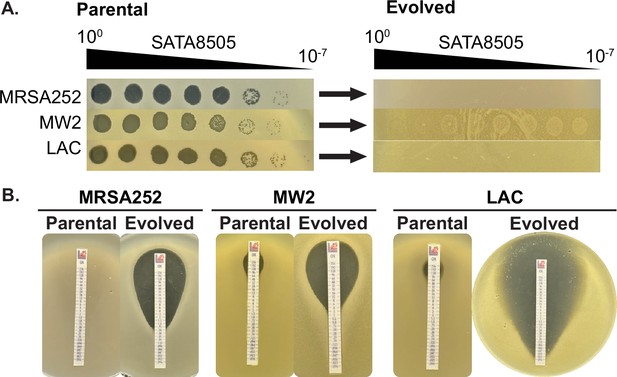
Phage SATA8505 infection drives loss of oxacillin resistance.
(A) MRSA strains MRSA252, MW2, and LAC treated with SATA8585 evolve resistance against SATA8585, evidenced by the reduction of plaquing from the parental to the evolved populations. (B) Evolved and parental MRSA show reduced MICs against oxacillin.
-
Figure 2—figure supplement 4—source data 1
Uncropped plate images for Figure 2—figure supplement 4A and B.
- https://cdn.elifesciences.org/articles/102743/elife-102743-fig2-figsupp4-data1-v1.zip
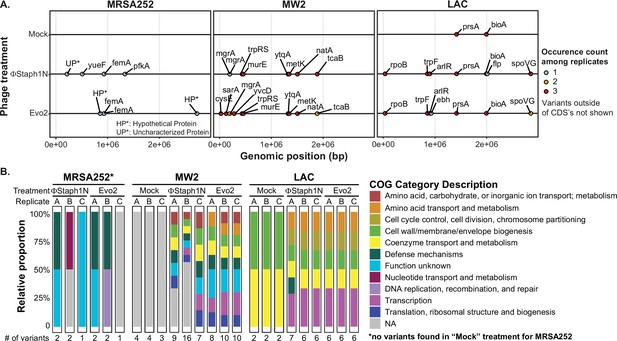
Phage infection of MRSA strains produces distinct mutational profiles.
(A) Coding sequences (CDS) with mutations from the three MRSA strains following phage treatment or mock treatment. For each strain, three isolates were sequenced and their mutations identified. Mutations are color-coded based on the number of occurrences among the three replicates. Information on all detected genetic variants is listed in Supplementary file 1. (B) Categories of genes with mutations that arose in each MRSA strain and treatment condition.
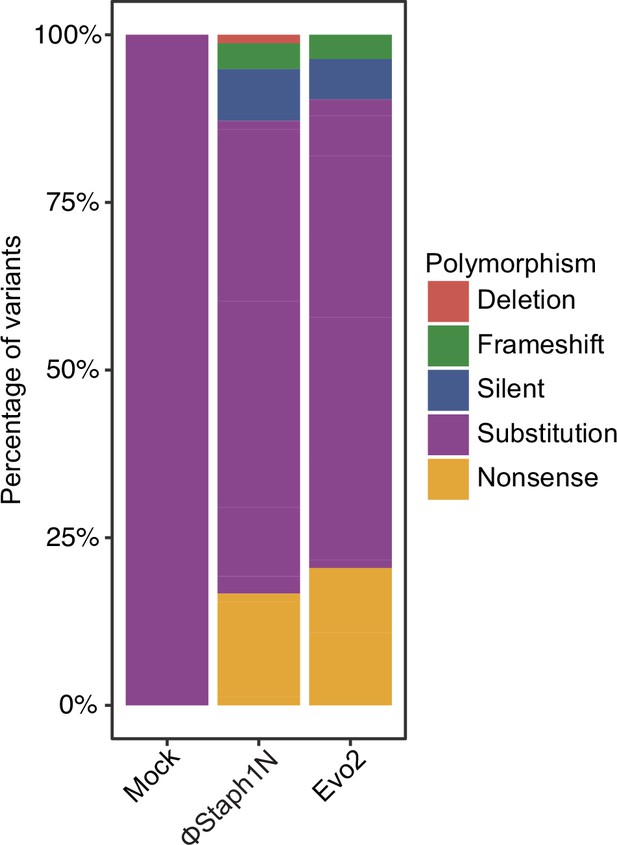
Types of polymorphisms in MRSA strains following infection by phage or a mock treatment.
Plotted are the polymorphisms that were found in a gene with an assigned COG category.
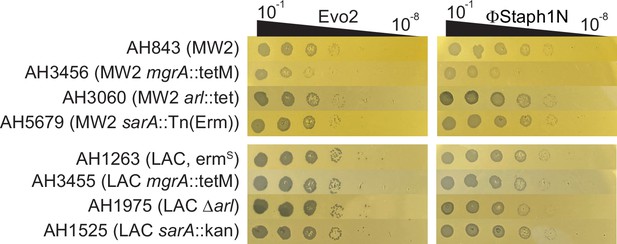
Plaquing efficiency of Evo2 and ΦStaph1N on MW2 and LAC strains with knockouts in mgrA, arl, and sarA.
-
Figure 3—figure supplement 2—source data 1
Uncropped plaquing images of Figure 3—figure supplement 2.
- https://cdn.elifesciences.org/articles/102743/elife-102743-fig3-figsupp2-data1-v1.zip
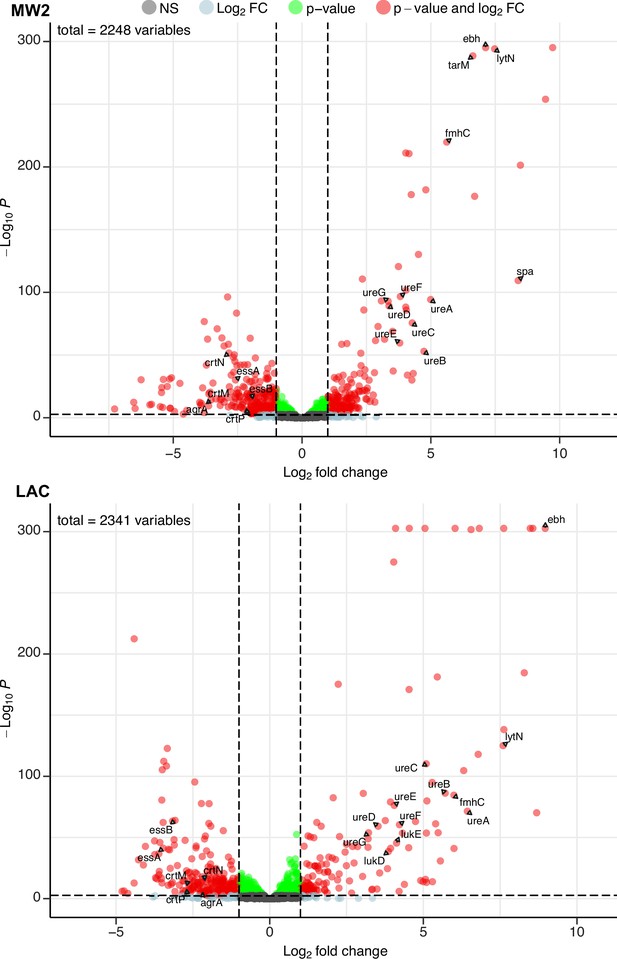
Phage infection changes the transcriptomic profile of MRSA.
Differential expression analysis was performed on the transcriptomes of MW2 (top panel) and LAC (bottom panel). For both strains, Evo2-infected samples were compared to uninfected controls. Three biological replicates were analyzed for each condition. Horizontal dotted lines represent an adjusted p-value cut-off of 0.002, while vertical dotted lines represent a log2 fold change of –2 or 2 in expression. Transcripts with a log2 fold change between –2 or 2 and a pvalue >0.002 are labeled as grey dots (Not significant, NS); transcripts that pass either the fold change or p-value cutoff (but not the other) are represented as blue and green dots, respectively; transcripts that pass both cutoffs are shown as red dots. Genes discussed in the main text are labeled. Data for all the transcripts with significant fold changes is shown in Supplementary file 2.
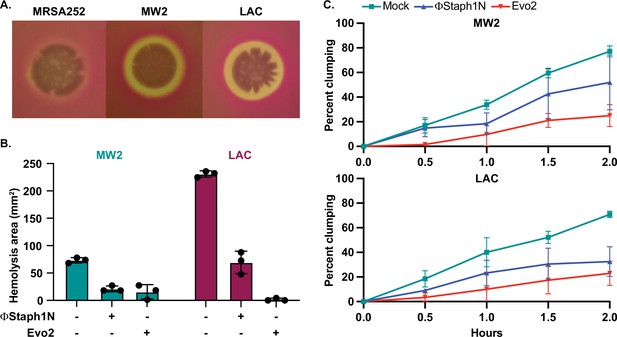
Phage treatment of MRSA results in attenuated virulence phenotypes.
(A) MW2 and LAC strains display hemolytic activity on rabbit blood agar plates, while MRSA252 does not. (B) Phage-treated MW2 and LAC strains display reduced hemolysis compared to uninfected cells. (C) Surviving cultures of MW2 and LAC treated with either ΦStaph1N (blue) or Evo2 (red) show reduced clumping rates compared to mock untreated cells (teal). Each condition was tested in three independent replicates and error bars represent the Standard Deviation (SD).
-
Figure 5—source data 1
Uncropped plates images for Figure 5A.
- https://cdn.elifesciences.org/articles/102743/elife-102743-fig5-data1-v1.zip
-
Figure 5—source data 2
Source data for the bar graphs in Figure 5B.
- https://cdn.elifesciences.org/articles/102743/elife-102743-fig5-data2-v1.xlsx
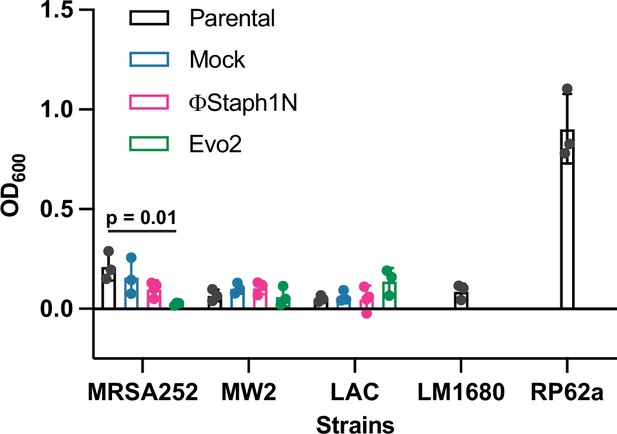
Effect of phage infection on biofilm formation in MRSA strains.
Cultures were infected or mock-infected with either ΦStaph1N or Evo2. RP62a is a strain of S. epidermidis with known biofilm-forming capability, while LM1680 is a derivative of RP62a that has lost biofilm-forming ability (Christensen et al., 1982; Jiang et al., 2013). Biofilm biomass was assessed by staining with Crystal Violet. Solubilized crystal violet was quantified by measuring absorbance at 600 nm. Values represent averages and Standard Deviations (SD) of three replicates. Statistical significance was determined with a two-tailed t-test.
-
Figure 5—figure supplement 1—source data 1
Source data for the bar graphs in Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/102743/elife-102743-fig5-figsupp1-data1-v1.xlsx
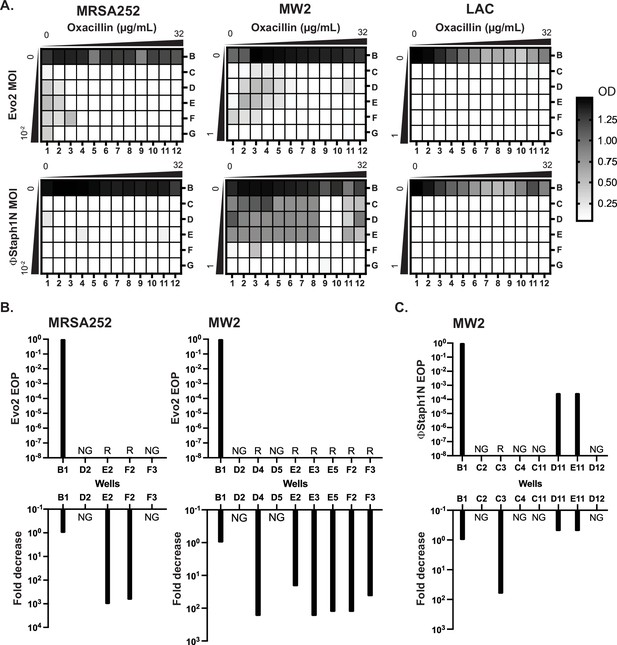
Co-treatment of MRSA with bacteriophage and β-lactam.
(A) Checkerboard assays of MRSA strains with gradients of oxacillin and Evo2 (top panels) or ΦStaph1N (bottom panels). The oxacillin gradient is a twofold serial dilution of drug concentration (µg/mL), while the phage MOI gradient is a 10-fold serial dilution of MOI. The rows and columns of each plate are labeled with letters and numbers, respectively. The black-white gradient in each well reflects the optical density of the culture and is the mean value from three biological replicates. MRSA strains co-treated with oxacillin and (B) Evo2 or (C) ΦStaph1N were tested for their phage resistance and oxacillin resistance. The letter/number combination reflects the well from which the cells were picked for analysis. Wells that could not produce a viable culture are labeled as NG (no growth). For wells that regrew, we calculated the efficiency of plaquing (EOP) of phage and measured the fold reduction in oxacillin MIC. Cultures that showed no detectable viral plaques are labeled as resistant (R).
Tables
Minimum inhibitory concentrations (µg/mL) against oxacillin of MRSA strains treated with different MOIs of phage.
| MRSA252 | ||||||
|---|---|---|---|---|---|---|
| ΦStaph1N | Evo2 | |||||
| MOI | Rep 1 | Rep 2 | Rep 3 | Rep 1 | Rep 2 | Rep 3 |
| 10–2 | 0.25 | 0.125 | 0.38 | NG | 2 | 0.5 |
| 10–3 | NG | 0.94 | 0.19 | 1 | 0.75 | 1 |
| 10–4 | 0.5 | 0.25 | 0.19 | 0.75 | 1 | 0.5 |
| 10–5 | 0.25 | 0.38 | NG | 0.38 | NG | NG |
| Mock | >256 | >256 | >256 | >256 | >256 | >256 |
| MW2 | ||||||
| ΦStaph1N | Evo2 | |||||
| MOI | Rep 1 | Rep 2 | Rep 3 | Rep 1 | Rep 2 | Rep 3 |
| 10–2 | 3 | 24 | 24 | 4 | NG | NG |
| 10–3 | 32 | 24 | 48 | 4 | NG | NG |
| 10–4 | 48 | 96 | 32 | 3 | NG | NG |
| 10–5 | 96 | 64 | 24 | 2 | NG | NG |
| Mock | 96 | 48 | 32 | 96 | 48 | 32 |
| LAC | ||||||
| ΦStaph1N | Evo2 | |||||
| MOI | Rep 1 | Rep 2 | Rep 3 | Rep 1 | Rep 2 | Rep 3 |
| 10–2 | NG | NG | 2 | 0.064 | NG | NG |
| 10–3 | NG | 3 | 1.5 | 0.032 | NG | NG |
| 10–4 | 32 | 1.5 | 1 | NG | NG | NG |
| 10–5 | 32 | 16 | 0.38 | NG | NG | NG |
| Mock | 32 | 48 | 48 | 32 | 48 | 48 |
-
NG: no growth detected.
Minimal inhibitory concentrations (µg/mL) of mock- or Evo2-treated MRSA strains against different antibiotics.
| Mock | Evo2 | ||||||
|---|---|---|---|---|---|---|---|
| Strain | Antibiotic | Rep 1 | Rep 2 | Rep 3 | Rep 1 | Rep 2 | Rep 3 |
| MRSA252 | Oxacillin | >256 | >256 | >256 | 0.38 | 0.75 | 0.5 |
| Rifampicin | 0.047 | 0.032 | 0.012 | 0.023 | 0.047 | 0.023 | |
| Mupirocin | 0.75 | 0.5 | 1 | 0.5 | 0.5 | 0.38 | |
| Erythromycin | >256 | >256 | >256 | >256 | >256 | >256 | |
| Teicoplanin | 6 | 6 | 4 | 4 | 4 | 0.75 | |
| Fosfomycin | 12 | 8 | 8 | 8 | 8 | 6 | |
| Daptomycin | 2 | 3 | 2 | 2 | 2 | 2 | |
| MW2 | Oxacillin | 48 | 32 | 32 | 0.75 | 1 | 0.75 |
| Rifampicin | 0.032 | 0.047 | 0.064 | 0.023 | 0.023 | 0.032 | |
| Mupirocin | 0.5 | 0.25 | 0.5 | 0.38 | 0.38 | 0.25 | |
| Erythromycin | 1 | 0.75 | 0.75 | 0.25 | 0.5 | 0.25 | |
| Teicoplanin | 2 | 1.5 | 1.5 | 1.5 | 1 | 1 | |
| Fosfomycin | 1 | 1.5 | 1.5 | 0.5 | 1 | 1 | |
| Daptomycin | 1.5 | 2 | 1.5 | 0.75 | 0.5 | 3 | |
| LAC | Oxacillin | 48 | 24 | 64 | 0.19 | 0.064 | 0.047 |
| Rifampicin | 0.047 | 0.047 | 0.047 | 0.032 | 0.032 | 0.047 | |
| Mupirocin | 0.5 | 0.75 | 0.75 | 0.5 | 0.5 | 0.5 | |
| Erythromycin | 3 | 3 | 3 | 1.5 | 1 | 2 | |
| Teicoplanin | 1 | 0.5 | 1 | 0.5 | 0.5 | 0.75 | |
| Fosfomycin | 6 | 6 | 12 | 1.5 | 12 | 1 | |
| Daptomycin | 0.5 | 3 | 1 | 0.064 | 2 | 0.75 | |
-
Rep = biological replicate.
Efficiencies of plaquing (EOPs)* of ΦStaph1N, Evo2, and ΦNM1γ6 on clinical isolates of USA300 (ADL1-30).
| Strain | ΦStaph1N | Evo2 | ΦNM1γ6 |
|---|---|---|---|
| RN4220 | 1.0E+00 | 1.0E+00 | 1.0E+00 |
| ADL1 | 1.7E-02 | 2.7E+00 | 6.7E-01 |
| ADL2 | 2.0E-01 | 3.3E+00 | 1.0E+00 |
| ADL3 | 6.0E-02 | 2.0E+00 | 1.7E-02 |
| ADL4 | 1.3E-03 | 1.0E+00 | 1.7E-01 |
| ADL5 | 1.2E-02 | 2.7E+00 | 1.0E+00 |
| ADL6 | 1.5E-02 | 1.7E+00 | 1.0E+00 |
| ADL7 | 6.0E-01 | 1.7E+00 | 6.7E-01 |
| ADL8 | 9.0E-02 | 2.0E+00 | 3.3E-01 |
| ADL9 | 6.0E-03 | 1.3E+00 | 2.0E-04 |
| ADL10 | 5.0E-01 | 1.3E+00 | 1.0E-03 |
| ADL11 | 1.0E+00 | 1.0E+00 | 3.3E-01 |
| ADL12 | 9.0E-04 | 2.0E+00 | 6.7E-07 |
| ADL13 | 7.0E-02 | 2.7E+00 | 2.3E-01 |
| ADL14 | 2.7E-01 | 3.3E+00 | 1.0E-05 |
| ADL15 | 9.0E-02 | 3.0E+00 | 2.0E-02 |
| ADL16 | 2.4E-01 | 5.6E+00 | 7.8E-02 |
| ADL17 | 3.3E+00 | 1.0E+01 | 2.4E+00 |
| ADL18 | 6.7E+00 | 1.4E+01 | 1.6E-02 |
| ADL19 | 7.1E-01 | 1.6E+00 | 2.3E+00 |
| ADL20 | 6.2E+00 | 7.8E+01 | 2.4E+00 |
| ADL21 | 5.2E+00 | 5.6E+00 | 3.3E-06 |
| ADL22 | 9.5E-01 | 2.0E+00 | 5.2E-01 |
| ADL23 | 1.5E+00 | 3.3E+01 | 1.2E-01 |
| ADL24 | 7.1E-01 | 5.6E+00 | 3.3E-06 |
| ADL25 | 7.1E-01 | 1.8E+00 | 2.3E-01 |
| ADL26 | 2.4E-02 | 3.9E+00 | 2.2E-03 |
| ADL27 | 1.3E+00 | 7.8E+00 | 1.7E+00 |
| ADL28 | 1.9E+00 | 8.9E+00 | 8.9E-02 |
| ADL29 | 1.9E+00 | 1.0E+01 | 1.3E-04 |
| ADL30 | 6.7E-01 | 1.4E+00 | 1.7E-05 |
-
*
Phage EOPs on the clinical isolates are standardized to their respective EOP on the laboratory strain S. aureus RN4220.
Mutated genes in MRSA following infection with phages ΦStaph1N or Evo2.
| Gene | Description | Strain | Phage infection/treatment | Reference |
|---|---|---|---|---|
| sarA | Transcriptional regulator of antibiotic resistance and virulence | MW2 | Evo2 | Li et al., 2016; Zielinska et al., 2012 |
| mgrA | Transcriptional regulator of antibiotic resistance and virulence | MW2 | ΦStaph1N, Evo2 | Crosby et al., 2016; Kwiecinski et al., 2021 |
| rpoB | Beta subunit of RNA polymerase Transcriptional regulator of antibiotic resistance | LAC | ΦStaph1N, Evo2 | Panchal et al., 2020 |
| arlR | Transcriptional regulator of antibiotic resistance and virulence | LAC | ΦStaph1N, Evo2 | Kwiecinski et al., 2021; Bai et al., 2019; Walker et al., 2013 |
| spoVG | Transcriptional regulator of antibiotic resistance and virulence | LAC | ΦStaph1N, Evo2 | Schulthess et al., 2011; Liu et al., 2016 |
| cysE | Cysteine and methionine synthesis, serine O-acetyltransferase | MW2 | Evo2 | Chen et al., 2019 |
| metK | Cysteine and methionine synthesis, S-adenosylmethionine (SAM) synthetase | MW2 | ΦStaph1N, Evo2 | Markham et al., 1984 |
| trpF | Phenylalanine, tyrosine and tryptophan synthesis, phosphoribosylanthranilate isomerase | LAC | ΦStaph1N, Evo2 | Proctor and Kloos, 1973 |
| femA | Peptidoglycan synthesis, pentaglycine synthesis | MRSA252 | ΦStaph1N, Evo2 | Maidhof et al., 1991; Srisuknimit et al., 2017 |
| murE | Peptidoglycan synthesis, UDP-MurNAc tripeptide synthesis | MW2 | ΦStaph1N, Evo2 | Gardete et al., 2004 |
| trpS | Aminoacyl-tRNA synthesis, tryptophanyl-tRNA synthesis | MW2 | ΦStaph1N, Evo2 | Xu et al., 1989 |
| ytqA | tRNA modifications, mnm5s2U synthesis | MW2 | ΦStaph1N, Evo2 | Jaroch et al., 2024 |
| yvcD | Unknown | MW2 | Evo2 | |
| natA | ABC transporter | MW2 | ΦStaph1N, Evo2 | Kobayashi et al., 2001 |
| tcaB | Predicted multidrug efflux pump | MW2 | ΦStaph1N, Evo2 | Maki et al., 2004 |
| fmhC | Fem-like factors | LAC | ΦΝΜ1γ6 | Willing et al., 2020 |
| rsaC ncRNA | modulates oxidative stress response and metal immunity | MW2 | ΦStaph1N+oxacillin | Lalaouna et al., 2019 |
| nrdF | class 1b ribonucleoside-diphosphate reductase subunit beta; beta subunit contains a metal-based cofactor; involved in DNA synthesis | MW2 | ΦStaph1N+oxacillin | Masalha et al., 2001 |
| fstAT ncRNA | Unknown | MW2 | ΦStaph1N+oxacillin | |
| rpoC | DNA-directed RNA polymerase subunit beta' | MW2 | ΦStaph1N+oxacillin | |
| tRNA | Transfer RNA | MW2 | ΦStaph1N+oxacillin |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Staphylococcus aureus) | MRSA MW2 (USA400) | Baba et al., 2002 | MW2 | |
| Strain, strain background (Staphylococcus aureus) | MRSA LAC (USA300) | Voyich et al., 2005 | LAC | |
| Strain, strain background (Staphylococcus aureus) | MRSA252 (USA200) | Holden et al., 2004 | MRSA252 | |
| Strain, strain background (Staphylococcus epidermidis) | S. epidermidis RP62a | Christensen et al., 1982 | RP62a | methicillin-resistant biofilm-producing S. epidermidis |
| Strain, strain background (Staphylococcus epidermidis) | S. epidermidis LM1680 | Jiang et al., 2013 | LM1680 | Derived from S. epidermidis RP62a; carries genomic deletion that inactivates biofilm production |
| Other | ΦStaph1N | Łobocka et al., 2012 | ΦStaph1N | Bacteriophage of the Kayvirus genus |
| Other | Evo2 | This study | Evo2 | Derived from ΦStaph1N |
| Other | ΦNM1γ6 | Marraffini laboratory | ΦNM1γ6 | Bacteriophage of the Dubowvirus genus, lytic version of temperate phage ΦNM1 |
| Other | ΦNM4γ4 | Marraffini laboratory | ΦNM4γ4 | Bacteriophage of the Dubowvirus genus, lytic version of temperate phage ΦNM4 |
| Other | Φ12 | Marraffini laboratory | Φ12 | Bacteriophage of the Triavirus genus |
| Other | Andhra | Hatoum-Aslan laboratory | Andhra | Bacteriophage of the Andravirus genus, infects S. epidermidis |
| Other | SATA8505 | Environmental isolate; Pincus et al., 2015 | SATA8505 | Bacteriophage of the Kayvirus genus, isolated from the environment in this study |
| Strain, strain background (Staphylococcus aureus) | ADL1 | Levin laboratory; Land et al., 2015 | ADL1 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL2 | Levin laboratory; Land et al., 2015 | ADL2 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL3 | Levin laboratory; Land et al., 2015 | ADL3 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL4 | Levin laboratory; Land et al., 2015 | ADL4 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL5 | Levin laboratory; Land et al., 2015 | ADL5 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL6 | Levin laboratory; Land et al., 2015 | ADL6 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL7 | Levin laboratory; Land et al., 2015 | ADL7 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL8 | Levin laboratory; Land et al., 2015 | ADL8 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL9 | Levin laboratory; Land et al., 2015 | ADL9 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL10 | Levin laboratory; Land et al., 2015 | ADL10 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL11 | Levin laboratory; Land et al., 2015 | ADL11 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL12 | Levin laboratory; Land et al., 2015 | ADL12 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL13 | Levin laboratory; Land et al., 2015 | ADL13 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL14 | Levin laboratory; Land et al., 2015 | ADL14 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL15 | Levin laboratory; Land et al., 2015 | ADL15 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL16 | Land et al., 2015 | ADL16 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL17 | Levin laboratory; Land et al., 2015 | ADL17 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL18 | Levin laboratory; Land et al., 2015 | ADL18 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL19 | Levin laboratory; Land et al., 2015 | ADL19 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL20 | Levin laboratory; Land et al., 2015 | ADL20 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL21 | Levin laboratory; Land et al., 2015 | ADL21 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL22 | Levin laboratory; Land et al., 2015 | ADL22 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL23 | Levin laboratory; Land et al., 2015 | ADL23 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL24 | Levin laboratory; Land et al., 2015 | ADL24 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL25 | Levin laboratory; Land et al., 2015 | ADL25 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL26 | Levin laboratory; Land et al., 2015 | ADL26 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL27 | Levin laboratory; Land et al., 2015 | ADL27 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL28 | Levin laboratory; Land et al., 2015 | ADL28 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL29 | Levin laboratory; Land et al., 2015 | ADL29 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | ADL30 | Levin laboratory; Land et al., 2015 | ADL30 | USA300, Patient isolate |
| Strain, strain background (Staphylococcus aureus) | AH1263 | Horswill laboratory | AH1263 | LAC, ErmS |
| Strain, strain background (Staphylococcus aureus) | AH3455 | Horswill laboratory | AH3455 | LAC mgrA::tetM |
| Strain, strain background (Staphylococcus aureus) | AH1975 | Horswill laboratory | AH1975 | LAC Δarl |
| Strain, strain background (Staphylococcus aureus) | AH1525 | Horswill laboratory | AH1525 | LAC sarA::kan |
| Strain, strain background (Staphylococcus aureus) | AH843 | Horswill laboratory | AH843 | MW2 |
| Strain, strain background (Staphylococcus aureus) | AH3456 | Horswill laboratory | AH3456 | MW2 mgrA::tetM |
| Strain, strain background (Staphylococcus aureus) | AH3060 | Horswill laboratory | AH3060 | MW2 arl::tet |
| Strain, strain background (Staphylococcus aureus) | AH5679 | Horswill laboratory | AH5679 | MW2 sarA::Tn(Erm) |
| Software, algorithm | Filtlong | Wick, 2021 | v.0.2.1; RRID:SCR_024020 | |
| Software, algorithm | Minimap2 | Li, 2018 | v.2.22; RRID:SCR_018550 | |
| Software, algorithm | SAMtools | Li et al., 2009 | v.13; RRID:SCR_002105 | |
| Software, algorithm | Bakta | Schwengers et al., 2021 | v.1.10.3; RRID:SCR_026400 | |
| Software, algorithm | eggNOG-mapper | Cantalapiedra et al., 2021 | v.2.1.12; RRID:SCR_021165 | |
| Software, algorithm | Bowtie 2 | Langmead and Salzberg, 2012 | v2.5.4; RRID:SCR_016368 | |
| Software, algorithm | featureCounts | Liao et al., 2014 | v.2.0.8; RRID:SCR_012919 | |
| Software, algorithm | R Project for Statistical Computing | https://www.r-project.org/ | v.4.4.0; RRID:SCR_001905 | |
| Software, algorithm | DESeq2 | Love et al., 2014 | v.1.44.0; RRID:SCR_015687 | |
| Software, algorithm | tidyverse | Wickham et al., 2019 | v.2.0.0; RRID:SCR_019186 | |
| Software, algorithm | EnhancedVolcano | https://github.com/kevinblighe/EnhancedVolcano | v.1.22.0; RRID:SCR_018931 | |
| Software, algorithm | Trimmomatic | Bolger et al., 2014 | v.0.39; RRID:SCR_011848 | |
| Software, algorithm | SPAdes | Bankevich et al., 2012 | v.4.0.0; RRID:SCR_000131 | |
| Software, algorithm | blastn | Altschul et al., 1997 | v2.16.0; RRID:SCR_001598 | |
| Software, algorithm | checkv | Nayfach et al., 2021 | v.1.0.3 | |
| Software, algorithm | taxmyphage | Millard et al., 2025 | v.0.3.4 | |
| Software, algorithm | Flye | Kolmogorov et al., 2019 | v.2.9.3; RRID:SCR_017016 | |
| Software, algorithm | Prodigal | Hyatt et al., 2010 | v.2.6.3; RRID:SCR_011936 |
Additional files
-
Supplementary file 1
List of mutations found in phage-treated and untreated S. aureus strains.
- https://cdn.elifesciences.org/articles/102743/elife-102743-supp1-v1.xlsx
-
Supplementary file 2
List of significantly up-or downregulated transcripts in MRSA strains MW2 and LAC.
- https://cdn.elifesciences.org/articles/102743/elife-102743-supp2-v1.xlsx
-
Supplementary file 3
Spanish translation of this article.
- https://cdn.elifesciences.org/articles/102743/elife-102743-supp3-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/102743/elife-102743-mdarchecklist1-v1.docx





