Yeast Eps15-like endocytic protein Pan1p regulates the interaction between endocytic vesicles, endosomes and the actin cytoskeleton
Figures
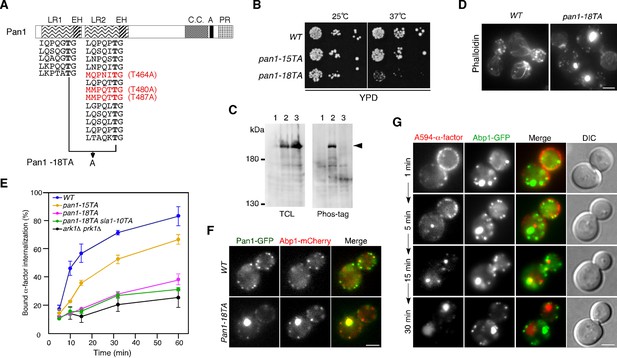
Construction and characterization of a Pan1p phosphorylation-site mutant.
(A) Structure of a Pan1p phosphorylation mutant. The two amino-terminal Eps15 homology (EH) domains, long repeat (LR) regions, predicted coiled-coil (CC), acidic region (A), and carboxy-terminal prolin-rich domain (PR) domain, are indicated. The fifteen consensus phosphorylation sites previously mutated in Pan1-15TA are indicated below the protein in black. The three additional sites mutated in Pan1-18TA are in red. (B) Plate showing the growth phenotype of pan1-15TA and pan1-18TA mutants. A dilution series of cells was plated on YPD plates and incubated for 2–3 days at 25 or 37°C, respectively. (C) Analysis of the phosphorylation state of the Pan1-18TA mutant. Protein expression was analyzed by immunoblotting 20 μg of total cell lysate (TCL) with an anti-GFP antibody (left panel). Phosphorylated proteins were purified from TCL with Phos-tag agarose, run on SDS-PAGE and immunoblotted with the anti-GFP antibody (right panel) as described in Materials and methods. Lane 1, JJTY369; lane 2, JJTY509; lane 3, JJTY5486. (D) Alexa Fluor 488-phalloidin staining of fixed wild-type and pan1-18TA cells to visualize actin. (E) The effect of Pan1p phosphorylation-site mutations on endocytic internalization. Radiolabeled α-factor internalization assays performed on wild-type (blue), pan1-15TA (yellow), pan1-18TA (magenta), pan1-18TA sla1-10TA (green), or ark1△ prk1△ (black) cells at 25°C. Each curve represents the average of three independent experiments, and error bars indicate the SD at each time point. (F) The localization of Pan1-GFP in wild-type and pan1-18TA cells. Cells expressing Pan1-GFP and Abp1-mCherry were grown to early to mid-logarithmic phase in YPD medium at 25°C and observed by fluorescence microscopy. Merged images of GFP and mCherry channels are shown in the right panels. (G) Endocytic cargo is transported to the vacuole through the actin clumps in pan1-18TA. pan1-18TA cells were labeled with A594-α-factor as described in the Methods. The images were acquired simultaneously at 1, 5, 15, and 30 min after washing out unbound A594-α-factor and warming the cells to 25°C. Scale bars, 2.5 μm.
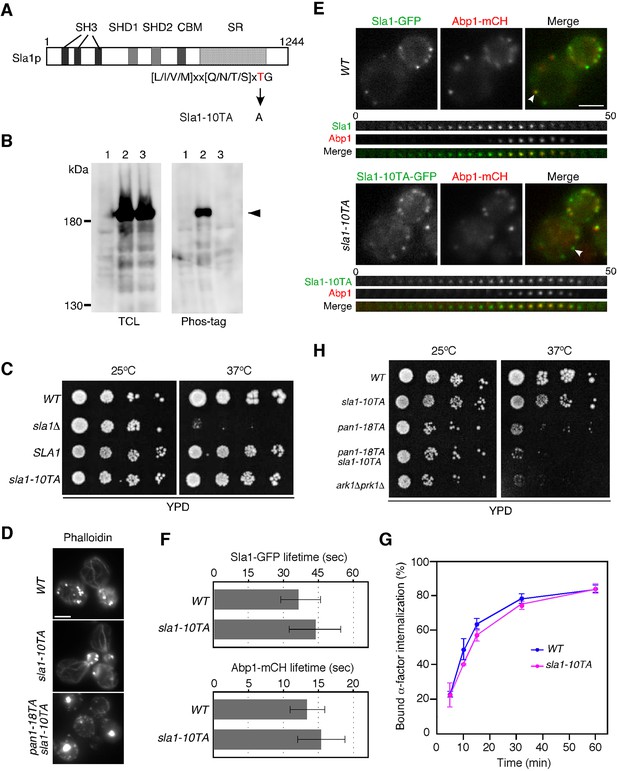
Construction and characterization of a Sla1p phosphorylation-site mutant.
(A) Structure of the Sla1p phosphorylation mutant. The three src homology 3 (SH3) domains, two Sla1 homology domains (SHD1/2), clathrin-binding motif (CBM) and carboxyl-terminal Sla1 repeats (SR) are indicated. The threonine in the 10 consensus Prk1 phosphorylation sites indicated below the protein was mutated to an alanine to make Sla1-10TA. (B) Phosphorylation state of Sla1-10TA mutant. Expression of proteins were analyzed by immunoblotting 20 μg total cell lysate (TCL) with anti-GFP antibody (left panel). Phosphorylated proteins were purified from TCL by Phos-tag agarose, run on SDS-PAGE and immunoblotted with anti-GFP antibody (right panel). Lane 1, JJTY369; lane 2, JJTY130; lane 3, JTY4139. (C) Plate showing the growth phenotype of the sla1-10TA mutant. A dilution series of cells was plated on YPD plates and incubated for 2–3 days at 25 or 37°C, respectively. (D) The in vivo effect of the Sla1p phosphorylation-site mutant. Wild-type, sla1-10TA, and pan1-18TA sla1-10TA cells were fixed and stained with Alexa Fluor 488-phalloidin to visualize actin. (E) Upper panels are single frames from a two-color movie showing Sla1-GFP (green) and Abp1-mCherry (red) in wild-type (top) or sla1-10TA (bottom) cells. Lower panels are time series of patches marked by arrowheads in upper panels. The time to acquire one image pair was 1 s. (F) Average lifetime of Sla1-GFP and Abp1-mCherry ± SD in wild-type and sla1-10TA cells. Data were taken from 60–90 s movies with a 1 sec frame interval. n = 50 patches for each strain. (G) Effect of Sla1p phosphorylation-site mutation on endocytic internalization. Radiolabeled α-factor internalization assays performed on wild-type (blue) or sla1-10TA (magenta) cells at 25°C. Each curve represents the average of three independent experiments, and error bars indicate the SD at each time point. Scale bars, 2.5 μm. (H) The growth phenotype of sla1-10TA, pan1-18TA, sla1-10TA pan1-18TA double mutants, and ark1△ prk1△. A dilution series of cells was plated on YPD plates and incubated for 2–3 days at 25 or 37°C, respectively.
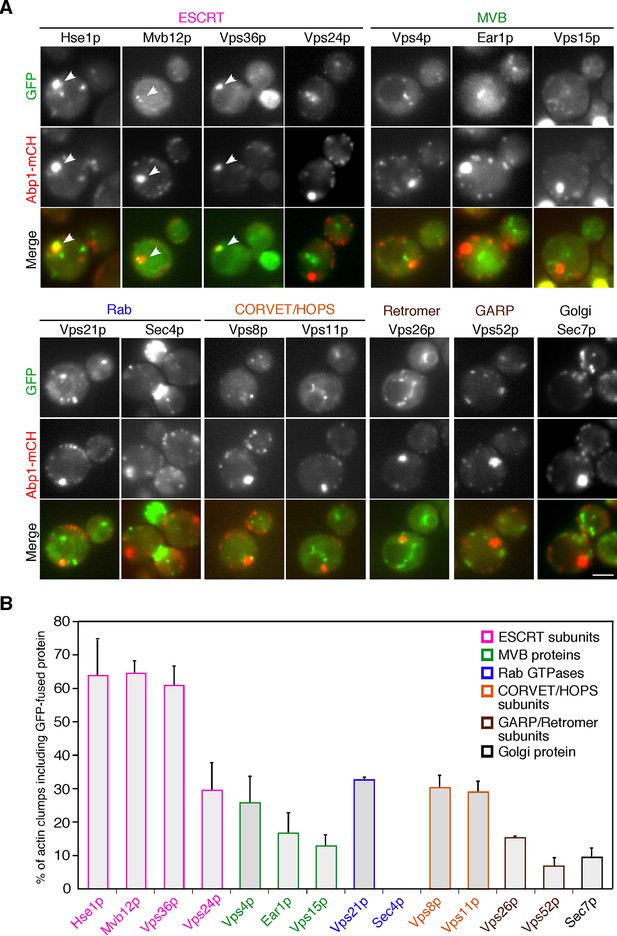
The localization of endosomal proteins in pan1-18TA cells.
(A) Localization of GFP-tagged endosomal proteins in pan1-18TA. pan1-18TA cells expressing Abp1-mCherry and GFP-tagged endosomal proteins were grown to early to mid-logarithmic phase in YPD medium at 25°C and observed by fluorescence microscopy. Merged images of GFP and mCherry channels are shown in the lower panel. Arrowheads indicate examples of colocalization. Scale bar, 2.5 μm. (B) Quantification of actin clumps including GFP-tagged endosomal proteins. The percentages were calculated as the ratio of actin clumps (n = 100) colocalizing with each protein in each experiment. Error bars indicate the standard deviation (SD) from at least three independent experiments.
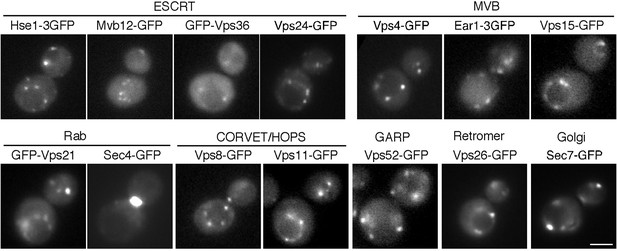
Localization of endosomal proteins in wild-type cells.
Cells expressing each GFP-tagged endosomal protein were grown to early to mid-logarithmic phase in YPD medium at 25°C and observed by fluorescence microscopy.
Scale bars, 2.5 μm.
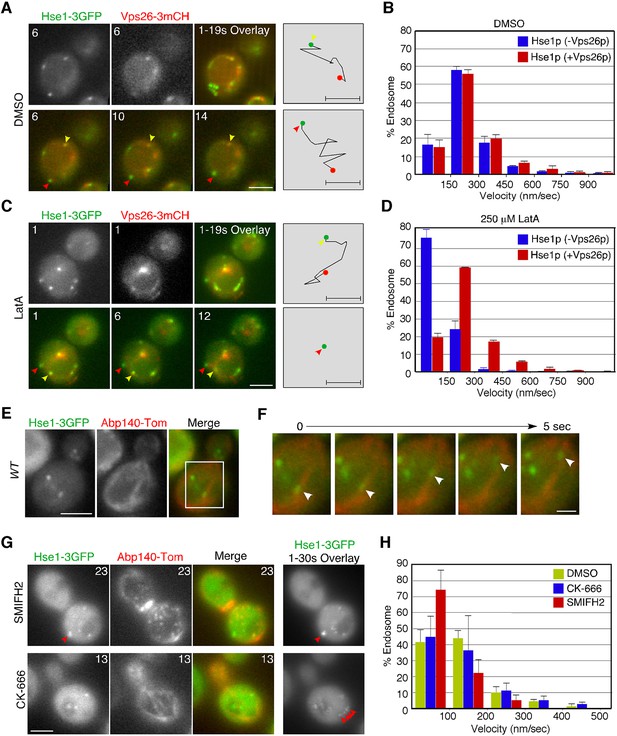
Interaction of endosomes expressing Hse1-3GFP with the actin cytoskeleton.
(A and C) Movement of Hse1-3GFP-containing endosomes in living cells. Wild-type cells expressing Vps26-3mCherry and Hse1-3GFP were grown to log phase at 25°C, treated with DMSO (LatA-) (A) or 250 μM LatA (C) for 30 min at 25°C, and subsequently imaged at 1 s intervals. In the upper row the left two panels show the individual channels and the right most image shows a merged overlay of the signal from both channels in the first 19 s. The lower row shows merged images of both channels at indicated time. Yellow or red arrowheads indicate examples of Hse1p-containing endosomes that do or do not co-label with Vps26p, respectively. Scale bars, 2.5 μm. The schematic panels on the right show tracking of the endosome indicated by yellow or red arrowheads in the lower microscope images. Scale bars, 0.5 μm. (B and D) Quantification of the velocity of Hse1p-containing endosomes. Endosome velocities were acquired at 1 s intervals and categorized according to a velocity range. (E) Hse1p-containing endosomes move along actin cables. Wild-type cells expressing Hse1-3GFP and Abp140-tdTomato were grown to early to mid-logarithmic phase and each image pair was acquired simultaneously at 1 s intervals. Scale bar, 2.5 μm. (F) Higher magnification view of the boxed area in (E) at successively later time points specified in s above. Arrowheads indicate an endosome moving along an actin cable. (G) The effect of SMIFH2 or CK-666 on movement of Hse1-3GFP-containing endosomes. Wild-type cells expressing Abp140-tdTomato and Hse1-3GFP were grown to log phase at 25°C, treated with 25 μM SMIFH2 (the upper row) or 100 μM CK-666 (the lower row) for 30 min at 25°C, and subsequently imaged at 1 s intervals. The left three panels show the individual channels and their merged image for a particular time point, and the right most image shows a merged overlay of Hse1-GFP signals in 30 sec. Red arrowheads indicate examples of Hse1p-containing endosomes. Scale bars, 2.5 μm. (H) Quantification of the velocity of Hse1p-containing endosomes. Endosome velocities were acquired at 1 s intervals and categorized according to a velocity range.
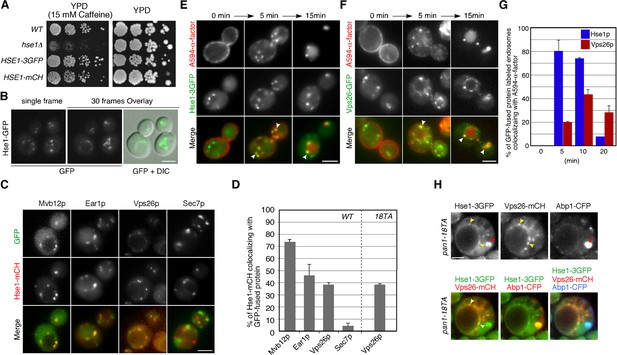
Localization of 3GFP- or mCherry-tagged Hse1p in wild-type cells.
(A) The functionality of 3GFP- and mCherry-tagged Hse1p was confirmed by testing their ability to complement the growth phenotype of hse1△ cells in a dilution series of each plated on YPD containing 15 mM caffeine, and incubated at 25°C. (B) Hse1p is localized at endosomes and PVCs in wild-type cells. The left panel is a single frame from a single-color movie showing Hse1-3GFP in wild-type cells. The middle panel is an image of the overlay of the Hse1-GFP localization seen in each 1 s frame interval of a 30 s movie. Examining Hse1-3GFP in the overlay of 30 consecutive time-lapse frames made it easy to distinguish the Hse1p localizing at endosomes in the cytoplasm and at the PVCs. The merged image of the Hse1-3GFP overlay and the differential interference contrast (DIC) images are shown in the right panel. (C) Localization of Hse1-mCherry and GFP-tagged proteins in living cells. Merged images of GFP and mCherry channels are shown in the lower panels. Wild-type cells expressing Hse1-mCherry and GFP-tagged proteins were grown to early to mid-logarithmic phase at 25°C in YPD medium and observed by fluorescence microscopy. Each image pair was acquired simultaneously. (D) The histogram represents the percentage of Hse1-mCherry labeled compartments colocalizing with the indicated organelle markers. In each experiment (n = 100) Hse1-mCherry labeled compartments were counted for each marker protein. Error bars indicate the SEM from at least three independent experiments. (E and F) Localization of 3GFP-tagged Hse1p (E) or Vps26p (F) in endocytic compartments. Cells were labeled with A594-α-factor as described in the Methods. The images were acquired simultaneously at 0, 5, and 15 min after washing out unbound A594-α-factor and warming the cells to 25°C. Arrowheads indicate examples of colocalization. (G) Quantification of the colocalization of Hse1-3GFP or Vps26-GFP with A594-α-factor at each time point. The percentages of colocalization were calculated as the ratio of A594-α-factor localized in the respective GFP positive compartments (n = 50) in each experiment. Error bars indicate the SEM from at least three independent experiments. (H) Hse1p resides both in actin clump and late endosomes. Each image pair was acquired using fluorescence microscopy equipped with high-speed filter changer. Time to acquire one image pair is 3.5 s. Arrowheads indicate example of colocalization of Hse1-3GFP and Vps26-mCherry (yellow) or Abp1-CFP (red). Scale bars, 2.5 μm.
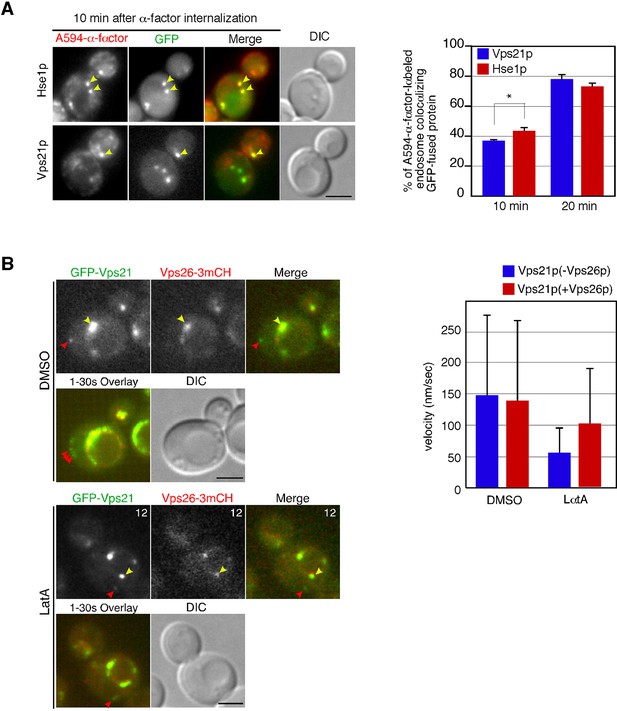
Localization and movement of GFP-Vps21-containing endosomes in living cells.
(A) Localization of Hse1-3GFP or GFP-Vps21 in endocytic compartments. Cells were labeled with A594-α-factor as described in the Materials and methods. The images were acquired simultaneously at 10 min after washing out unbound A594-α-factor and warming the cells to 25°C. Arrowheads indicate examples of colocalization. The bar graphs represent the colocalization of Hse1-3GFP or GFP-Vps21 with A594-α-factor at 10 or 20 min after A594-α-factor internalization. The percentages of colocalization were calculated as the ratio of GFP signals in A594-α-factor positive compartments (n = 50) in each experiment. Error bars indicate the SEM from at least three independent experiments. (B) Movement of GFP-Vps21-containing endosomes in living cells. Wild-type cells expressing Vps26-3mCherry and GFP-Vps21 were grown to log phase at 25°C, treated with DMSO (LatA-) or 250 μM LatA for 30 min at 25°C, and subsequently imaged at 1 s intervals. In the upper row the left two panels show the individual channels and the right most image shows a merged images. The lower left panel shows merged overlay of the signal from both channels in the first 30 s. The bar graphs represent the average velocity of Vps21p-containing endosomes (n = 100). Scale bars, 2.5 μm.
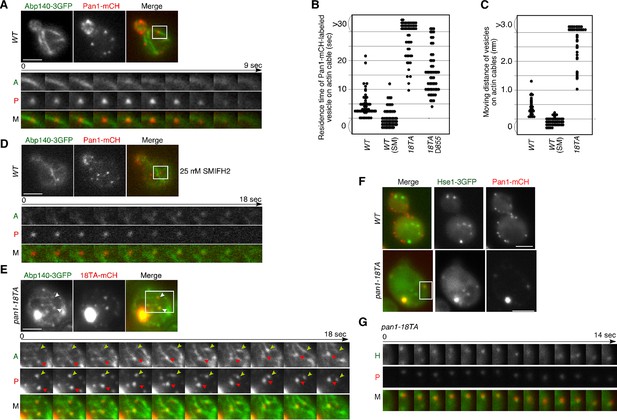
Interaction of Pan1p-containing compartments with actin cables and endosomes.
(A) Localization of Abp140-3GFP and Pan1-mCherry in a wild-type cell. The lower panels correspond to a time series of a higher magnification view of the boxed area in the upper right image. (B) The residence time of Pan1-mCherry-labeled vesicles on an actin cable. The residence time was determined from 60 sequential two-dimensional images. n = 52 vesicles for each strain. Vesicles residing on cable over 30 s are indicated as >30 in the graph. (C) Moving distance of Pan1-mCherry-labeled vesicles on actin cables. To determine each moving distance, the distance that the center of the Pan1-mCherry fluorescence moves on an actin cable was calculated based on pixel coordinates (1 pixel = 64.5 nm). n = 42 vesicles for each strain. (D) Effect of the formin inhibitor SMIFH2 on the movement of Pan1-mCherry patches. Wild-type cells expressing Pan1-mCherry and Abp140-3GFP were grown to log phase at 25°C, treated with 25 μM SMIFH2 for 30 min at 25°C, and subsequently imaged at 1 s intervals. (E) The localization of Abp140-3GFP and Pan1-18TA-mCherry in a pan1-18TA cell. The lower panels are single focal plane images corresponding to a time series of a higher magnification view of the boxed area in the upper right image. Arrowheads indicate examples of Pan1p-containing compartments moving along an actin cable. Yellow or red arrowheads indicates different vesicles. Upper and middle panels show GFP and mCherry channels, respectively, and lower panel shows their merged images. (F) The localization of Hse1-3GFP and Pan1-mCherry in wild-type and pan1-18TA cells. (G) Time series of single patches in the boxed area in (F). Cells expressing Hse1-3GFP and Pan1-18TA-mCherry were grown to early to mid-logarithmic phase at 25°C in YPD medium and imaged at 1 s intervals. Scale bars, 2.5 μm.
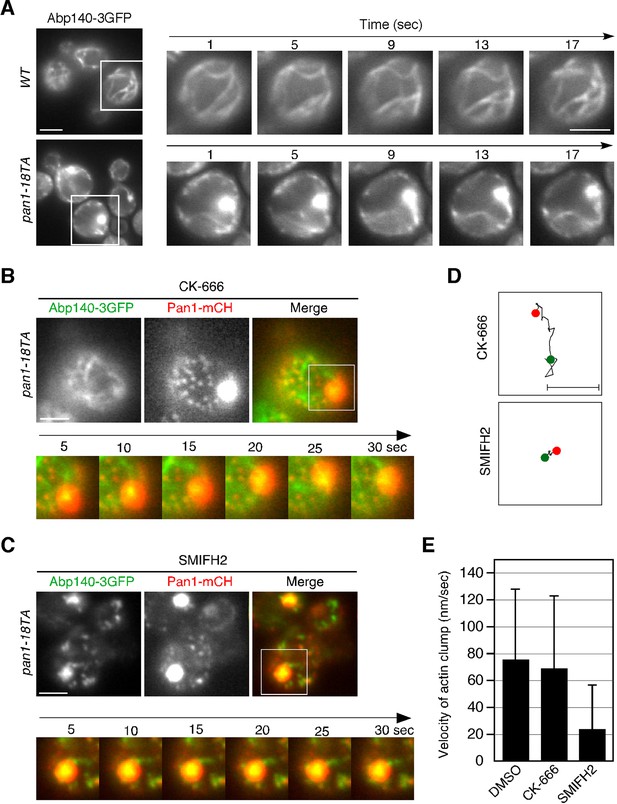
Actin cable dynamics in wild-type and pan1-18TA cells.
(A) Cells expressing Abp140-3GFP were grown to early to mid-logarithmic phase at 25°C in YPD medium and imaged at 1 s intervals. Right panels indicate a high magnification view of the boxed area in the left panels. (B and C) Effect of SMIFH2 or CK-666 on movement of actin clump. pan1-18TA cells expressing Abp140-3GFP and Pan1-mCherry were grown to log phase at 25°C, treated with 100 μM CK-666 (B) or 25 μM SMIFH2 (C) for 30 min at 25°C, and subsequently imaged at 1 s intervals. The upper images show the localization of Abp140-3GFP and Pan1-18TA-mCherry in a pan1-18TA cell. The lower panels correspond to a time series of a higher magnification view of the boxed area in the upper right image. (D) The schematic panels show tracking of the actin clumps in the boxed areas in (B) or (C). Positions of actin clumps were determined by calculating the center of fluorescence intensity. (E) The bar graphs represent the average velocity of actin clumps (n = 50). Scale bars, 0.5 μm.
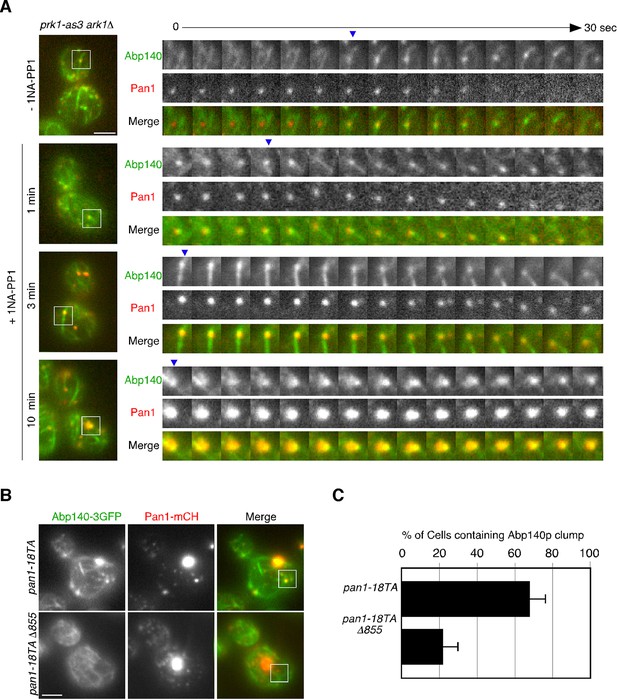
Interaction between endocytic vesicles and actin cables upon inhibition of Prk1p.
(A) The left images represent single frames from movies of ark1△ prk1-as3 mutant cells showing merged images of the GFP (Abp140p) and the mCherry (Pan1p) channel. The ark1△ prk1-as3 mutant cells expressing Abp140-3GFP and Pan1-mCherry were grown to log phase at 25°C, treated with 100 μM 1NA-PP1 for the indicated time at 25°C, and subsequently imaged at 1 s intervals. A time series of single patches in the boxed area for each strain are shown in the right panels. Blue arrowheads indicate Pan1-mCherry-labeled vesicles associating with actin cables. Scale bar, 2.5 μm. (B) Localization of Abp140-3GFP and Pan1-mCherry in pan1-18TA and pan1-18TA△855 cells. Scale bar, 2.5 μm. (C) Quantification of cells containing actin clumps. Cells expressing Abp140-3GFP were grown to log phase at 25°C and imaged. Data show mean ± SD from at least three experiments, with 50 cells counted for each strain per experiment.
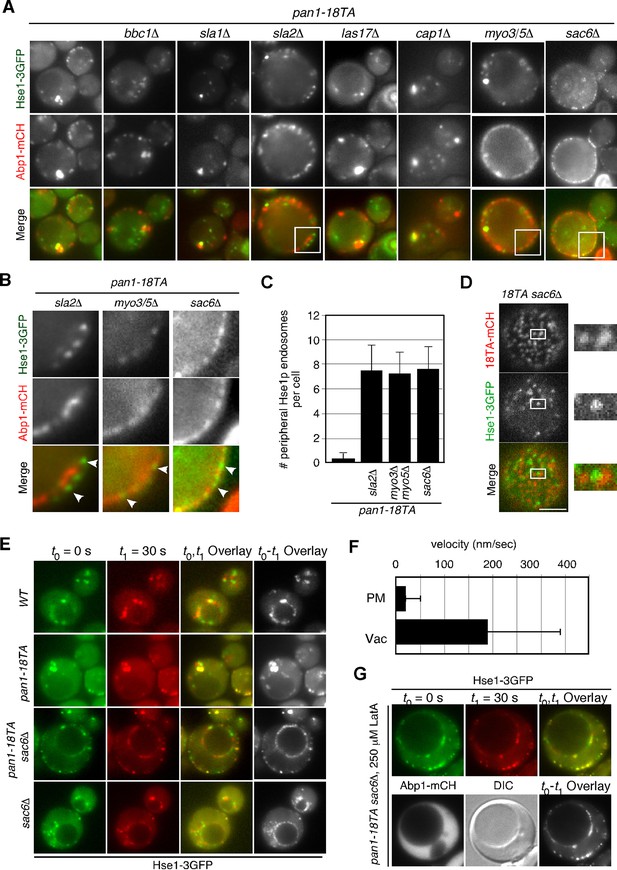
Interaction between endocytic vesicles and early endosomes in the pan1-18TA mutant.
(A) Localization of Hse1-3GFP-labeled endosomes and actin structures in pan1-18TA double mutant cells. The pan1-18TA and double mutant cells expressing Hse1-3GFP and Abp1-mCherry were grown to early to mid-logarithmic phase at 25°C in YPD medium and observed by fluorescence microscopy. (B) Higher magnification view of the boxed areas in (A). Arrowheads indicate examples of Hse1p-containing endosomes at the plasma membrane. (C) Quantification of the number of Hse1p-containing endosomes localizing at the cell periphery in single focal plane images. Data show mean ± SD, with n = 50 cells counted for each strain. (D) Localization of Pan1-18TA-mCherry and Hse1-3GFP in pan1-18TA sac6△ double mutant cells. (E) Movement of Hse1p-containing endosomes in pan1-18TA, sac6△, and the double mutant. Cells expressing Hse1-3GFP to visualize the endosomes were grown to log phase at 25°C, and imaged for 30 s at 1 s intervals. At time t0 = 0 s, Hse1-GFP is shown in green, at t1 = 30 s, Hse1-3GFP is shown in red. t0,t1 overlay shows overlay image of t0 and t1, and t0-t1 overlay shows overlay image of all 30 frames from t0 (0 s) to t1 (30 s). Scale bars, 2.5 μm. (F) Quantification of the velocity of Hse1p-containing endosomes at the cell periphery (PM) and the vacuolar membrane (Vac). (G) Localization of Hse1p-residing endosomes in the pan1-18TA sac6△ mutant treated with 250 μM LatA. Cells treated with 250 μM LatA for 30 min at 25°C were imaged for 30 s at 1 s intervals. At time t0 = 0 s, Hse1-GFP is shown in green, at t1 = 30 s, Hse1-3GFP is shown in red. t0,t1 overlay shows overlay image of t0 and t1. Scale bars, 2.5 μm.
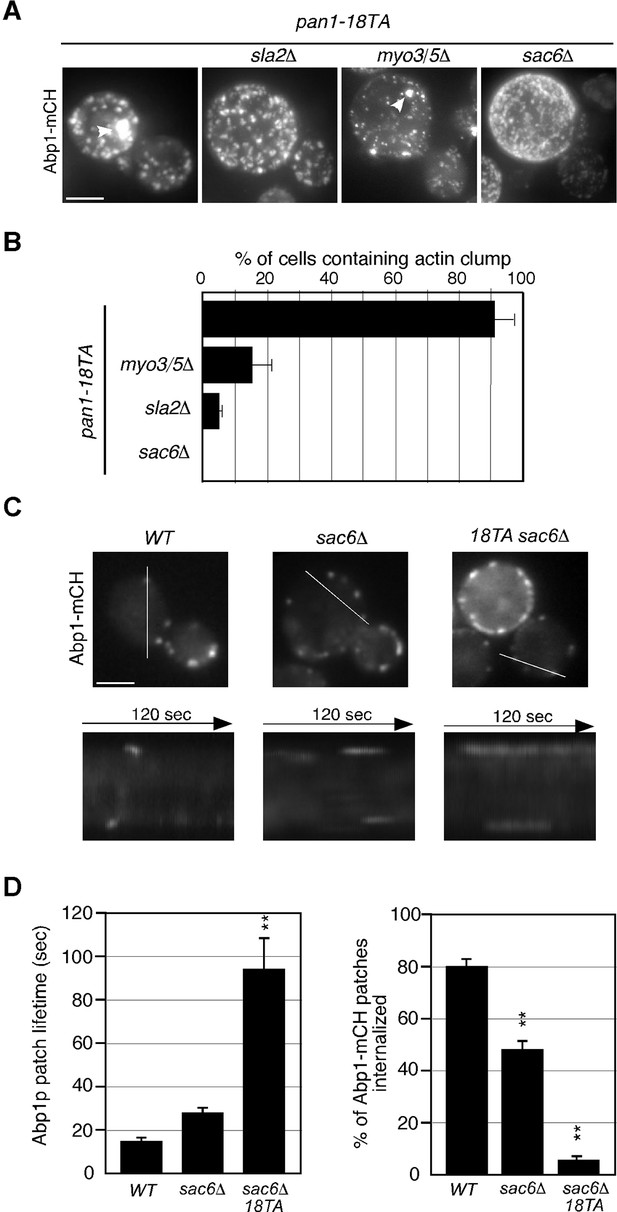
Actin structures and actin patch dynamics in pan1-18TA double mutant cells.
(A) Maximum intensity projections of Z stacks of pan1-18TA and double mutant cells labeled with Abp1-mCherry. The Z series was acquired through the entire cell at 0.2 μm intervals. (B) Quantification of the fraction of cells containing actin clumps. The bar graphs represent the average percentage of cells containing actin clump(s). The size of the actin clumps is not considered in this experiment. Data show mean ± SEM from at least three experiments, with 50 cells counted for each strain per experiment. (C) Localization of Abp1-GFP in wild-type, sac6△, and pan1-18TA sac6△ cells. Kymographs from the same movies are shown in the lower panels. (D) The left bar graph represents average lifetimes of Abp1-GFP ± SD in indicated cells. Data were taken from 1- or 2-min movies with a 1- or 2-s frame interval. n = 50 patches for each strain. **, p value <0.001, unpaired t-test. The right graph represents the percentage of patches internalized into the cytoplasm in indicated cells. Data show mean ± SEM from at least three experiments, with >50 patches counted for each strain per experiment. **, p value <0.001, unpaired t-test. Scale bars, 0.5 μm.
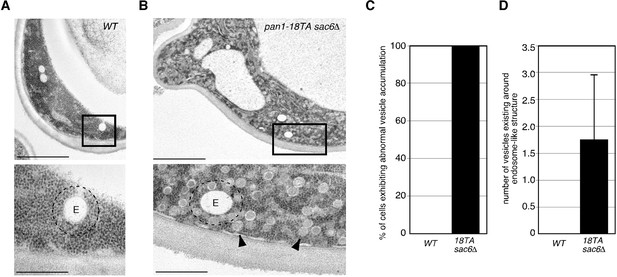
Ultrastructure of endocytic vesicles and endosomes observed in wild-type and pan1-18TA sac6△ cells.
(A and B) Wild-type and pan1-18TA sac6△ cells were grown at 25°C, fixed using propane jet freezing method and processed for electron microscopic analysis. The lower panels show higher magnification views of the boxed areas in the upper panels. Arrowheads point to endocytic vesicle-like structures that accumulate in pan1-18TA sac6△ cells. E: endosome-like structures. Dotted lines represent the areas 100 nm outside from endosome-like structures. Scale bars: 2 μm (upper panels), 0.5 μm (lower panels). (C) The bar graph represents the percentage of cell containing accumulation of ~50 nm vesicle (n = 20 cells). (D) Quantification of number of ~50 nm vesicles existing around endosome-like structures. Numbers of vesicles locating within 100 nm from outside of endosome-like structures were counted. Data show mean ± SD, with >50 endosomes counted for each strain.
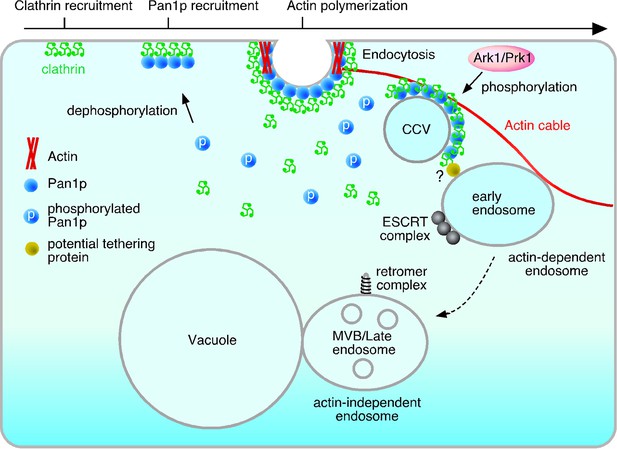
Model of the actin cable-mediated endocytic pathway.
Unphosphorylated Pan1p on an endocytic vesicle binds to actin to fix the vesicle to the actin cable. After being pinched off from the membrane, the endocytic vesicle moves into the cytosol as a result of actin cable flow, and then, interacts with the early endosome via potential tethering protein. Pan1p phosphorylation by Ark1/Prk1 kinases causes dissociation of coat proteins and the actin cable from the endocytic vesicle, making it possible for the vesicle to fuse to the endosome. This also results in the dissociation of the actin cable and the early endosome, which then moves to the vacuolar membrane, and matures into a late endosome.
Videos
Localization of Hse1-3GFP (left; green in merge) and Abp140-tdTomato (center; red in merge) in wild-type cells.
The interval between frames is 1 s.
Localization of Hse1-3GFP (left; green in merge) and Abp140-Tomato (center; red in merge) in wild-type cells treated with 25 μM SMIFH2 or 100 μM CK-666.
Arrowheads indicate examples of Hse1p-labeled endosome. The interval between frames is 1 s.
Localization of Abp140-3GFP in wild-type and pan1-18TA cells.
The interval between frames is 1 s.
Localization of Abp140-3GFP (left; green in merge) and Pan1-18TA-mCherry (center; red in merge) in pan1-18TA cells.
The interval between frames is 1 s.
Localization of Hse1-3GFP (left; green in merge) and Pan1-18TA-mCherry (center; red in merge) in pan1-18TA cells.
Arrowheads indicate examples of colocalization. The interval between frames is 1 s.
Localization of Abp140-3GFP (left; green in merge) and Pan1-mCherry (center; red in merge) in ark1△ prk1-as3 cells untreated or treated with 100 μM 1NA-PP1.
Arrowheads indicate examples of vesicles associated with actin cables. The interval between frames is 1 s.
Additional files
-
Supplementary file 1
Yeast strains used in this study.
- https://doi.org/10.7554/eLife.10276.023
-
Supplementary file 2
Primers used in this study.
- https://doi.org/10.7554/eLife.10276.024





