Ferritinophagy via NCOA4 is required for erythropoiesis and is regulated by iron dependent HERC2-mediated proteolysis
Figures
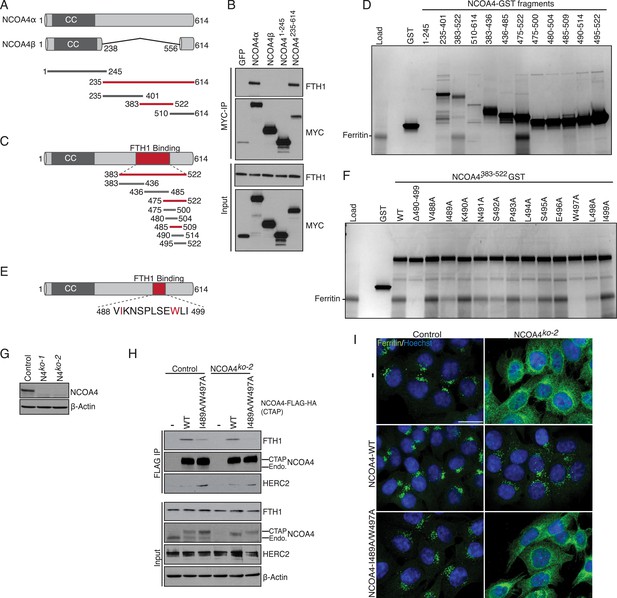
NCOA4 recognizes FTH1 via a conserved C-terminal domain.
(A) Schematic of NCOA4 truncation constructs. (B) Horse spleen apoferritin was mixed with in vitro translated MYC-NCOA4 fragments, immunoprecipitated with anti-MYC (MYC-IP) and immunoblotted with the indicated antibodies. (C) Schematic of NCOA4383−522 truncation constructs. (D) GST pulldown assay of recombinant NCOA4-GST fragments as indicated mixed with ferritin. Bound ferritin was analyzed by 4–20% SDS-PAGE and Coomassie blue staining. Load is 5% of ferritin input. (E) Schematic and sequence of NCOA4 point mutations designed within NCOA4383−522. (F) GST pulldown assay of recombinant NCOA4-GST fragments as indicated mixed with ferritin. Bound ferritin was analyzed by 4–20% SDS-PAGE and Coomassie blue staining. Load is 5% of ferritin input. (G) CRISPR/Cas9-mediated depletion of NCOA4 expression in HCT116 cells. Two independent NCOA4 knockout clones were generated following targeting of NCOA4 exon 2. A CRISPR non-targeting control cell line was also established. (H) Control or NCOA4ko−2 cells were transduced with NCOA4WT or NCOA4I489A/W497A lentivirus and stable cell lines were generated. Endogenous FTH1 binding was evaluated by immunoblot following FLAG affinity purification of WT or mutant NCOA4. (I) NCOA4I489A/W497A attenuates FTH1 localization in lysosomes following iron chelation. HCT116 control or NCOA4 knockout cells were plated on glass coverslips and treated with FAC for 14 hr. To promote ferritin accumulation in lysosomes, cells were then washed and treated with DFO plus lysosomal protease inhibitors E64-d and Pepstatin A for 6 hr. Cells were fixed, stained with ferritin antibody and visualized by confocal microscopy. Scale bar, 20 μm.
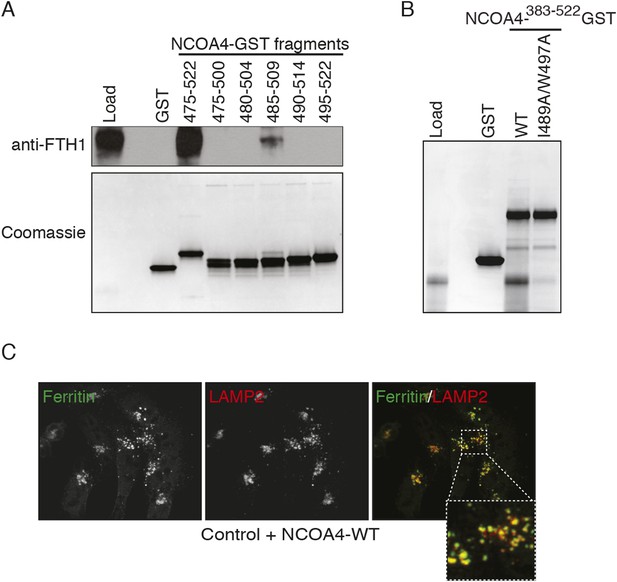
NCOA4 interacts with ferritin via a C-terminal domain and promotes lysosomal ferritin accumulation upon iron depletion.
(A) GST pulldown assay of recombinant NCOA4-GST fragments as indicated mixed with ferritin. Bound ferritin was analyzed by immunoblotting with FTH1 antibody. Load is 5% of ferritin input. NCOA4-GST fragment loading assessed by 4–20% SDS-PAGE followed by Coomassie blue stain. (B) GST pulldown assay of recombinant NCOA4-GST fragments as indicated mixed with ferritin. Bound ferritin was analyzed by 4–20% SDS-PAGE and Coomassie blue staining. Load is 5% of ferritin input. (C) FTH1 accumulates in lysosomes following iron chelation in NCOA4WT cells. HCT116 CRISPR control cells were plated on glass coverslips and treated with FAC for 14 hr; cells were then washed, treated with DFO plus lysosomal protease inhibitors E64-d and Pepstatin A for 6 hr, fixed and stained with FTH1 and LAMP2 specific antibodies. FTH1/LAMP2 localization was visualized by confocal microscopy.
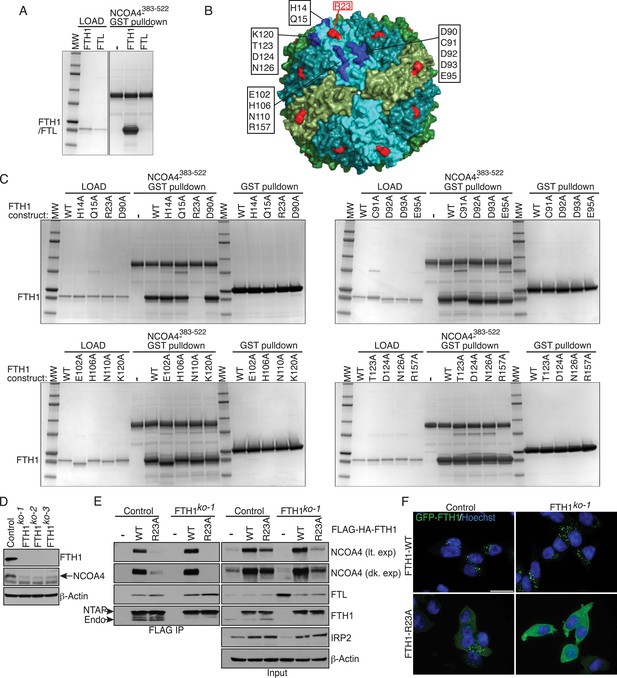
FTH1R23 is essential for NCOA4 binding and ferritinophagy.
(A) GST pulldown assay of recombinant NCOA4383−522-GST mixed with recombinant FTH1-only or FTL-only complexes. Bound FTH1 or FTL was analyzed by 4–20% SDS-PAGE and Coomassie blue staining. Load is 5% of FTH1 or FTL input. (B) Space-filling model of FTH1 ferritin complex (PDBid: 3AJO) with individual subunits colored in greens and blues, 16 conserved surface residues are highlighted in dark blue or red (R23) and labeled as indicated. (C) GST pulldown assay of recombinant NCOA4383−522-GST or GST alone mixed with recombinant wild type (WT) and point mutant (as indicated) FTH1-only complexes. Bound FTH1 or FTL was analyzed by 4–20% SDS-PAGE and Coomassie blue staining. Load is 5% of FTH1 input. (D) CRISPR/Cas9-mediated depletion of FTH1 expression in HCT116 cells. Three independent FTH1 knockout clones were generated following targeting of FTH1 exon 1. (E) Control or FTH1ko−1 cells were transduced with FTH1WT or FTH1R23A lentivirus and stable cell lines were generated. Endogenous NCOA4 and FTL binding was evaluated by immunoblot following FLAG affinity purification of WT or mutant FTH1. (F) Mutation of FTH1R23 attenuates GFP-FTH1 localization in lysosomes. HCT116 control or FTH1 knockout cells were plated on glass coverslips and treated with Bafilomycin for 6 hr to prevent lysosomal degradation of ferritin. Cells were fixed and visualized for GFP-FTH1 localization by confocal microscopy. Scale bar, 20 μm.
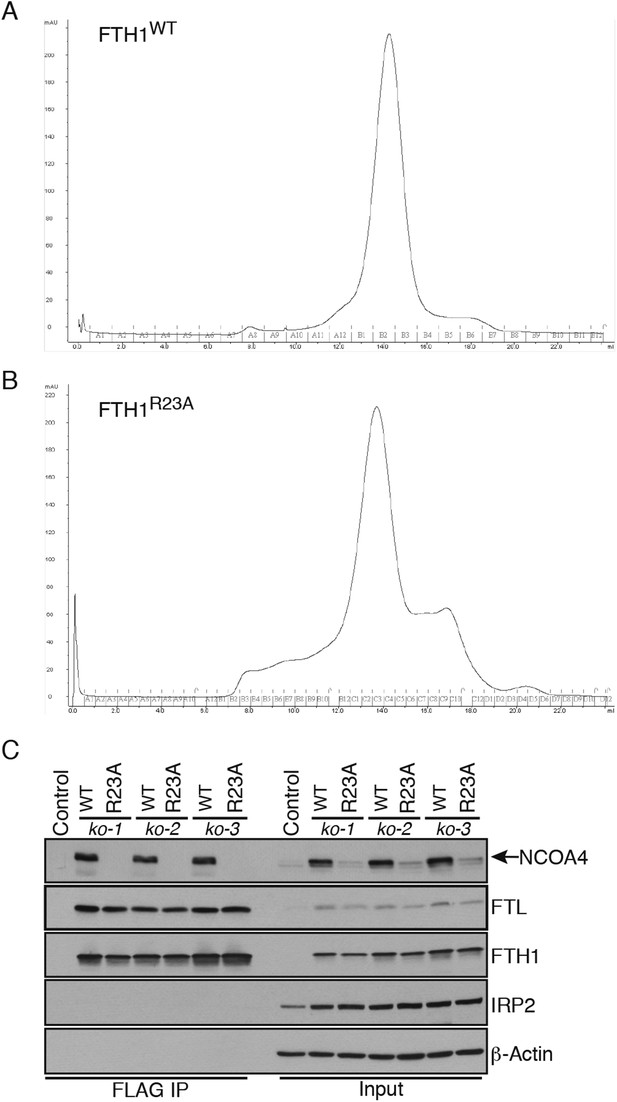
FTH1 R23A forms functional ferritin cages and abrogates NCOA4 binding.
(A) Size exclusion chromatogram of recombinant wild-type FTH1-only complexes with main peak noted at approximately 14 ml corresponding to an apparent 450 kDa molecular weight. (B) Size exclusion chromatogram of recombinant R23A FTH1-only complexes with main peak noted at approximately 14 ml corresponding to an apparent 450 kDa molecular weight. (C) Control or FTH1-deficient cells (three independent CRISPR clones) were transduced with FTH1WT or FTH1R23A lentivirus and stable cell lines were generated. Endogenous NCOA4 and FTL binding was evaluated by immunoblot following FLAG-IP of WT or mutant FTH.
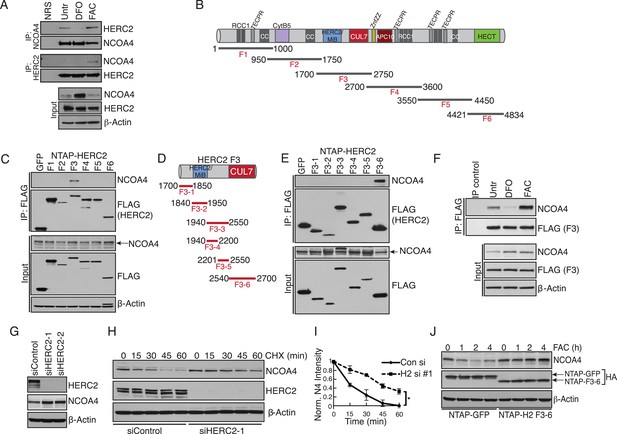
Iron-dependent NCOA4 turnover via the HERC2 E3 Ubiquitin Ligase.
(A) Endogenous IP of NCOA4 from 293T cells treated with DFO or FAC for 6 hr. Lysate input for control IgG IP is the untreated sample. HERC2 and NCOA4 binding was evaluated by immunoblot. (B) Schematic of HERC2 functional domains. To interrogate NCOA4 binding to HERC2, six fragments spanning the HERC2 protein (F1-6) were subcloned and expressed in 293T cells. (C) Endogenous NCOA4 binding to NTAP-HERC2 fragments. Extracts from 293T cells stably expressing the indicated proteins were immunoprecipitated with anti-Flag (Flag-IP) and immunoblotted with the indicated antibodies. (D) Identification of the minimal HERC2 domain responsible for NCOA4 binding. HERC2F3 was divided into six sub-fragments as indicated. (E) NCOA4 binds the CUL7 (CPH) homology domain of HERC2. HERC2 FLAG-tagged fragments 3-1 through 3–6 were expressed in 293T cells followed by Flag-IP, and immunoblot with the indicated antibodies. (F) HERC2 CUL7 domain binds endogenous NCOA4 in an iron-dependent manner. FLAG-tagged HERC2F3 was expressed in 293T cells, followed by DFO or FAC treatment for 9 hr and Flag-IP as in (C). Lysate input for the control IP (normal mouse serum) corresponds to the untreated sample. (G) siRNA-mediated knockdown of HERC2 expression promotes NCOA4 protein accumulation. U2OS cells were transfected with control or HERC2-specific siRNAs for 72 hr, harvested, and subjected to SDS-PAGE and immunoblot with the indicated antibodies to evaluate HERC2 knockdown and NCOA4 abundance. (H) NCOA4 half-life is extended upon depletion of HERC2. Immunoblot of NCOA4 protein levels in U2OS cells following control or HERC2 siRNA delivery and cycloheximide treatment (CHX) as indicated. (I) Quantification of NCOA4 protein levels in 2 independent biological CHX experiments. Error bars represent +/− standard deviation, *, p < 0.05 by two-tailed unpaired t-test at each time point. (J) Expression of HERC22540−2700 abrogates iron-mediated NCOA4 downregulation. 293T cells were transfected with FLAG-tagged HERC2F3−6 or FLAG-tagged GFP as a control, followed by FAC treatment as indicated. NCOA4 protein level was evaluated by immunoblot.
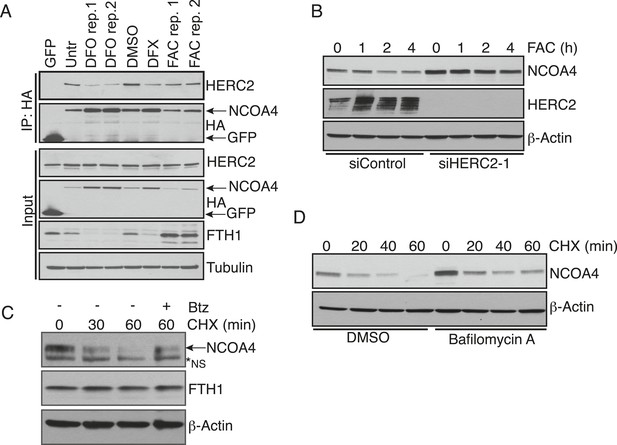
HERC2 regulates NCOA4 turnover.
(A) FLAG affinity purification of stably expressed NCOA4-FLAG-HA (CTAP) from 293T cells treated as indicated for 9 hr (100 μM DFO, 30 μM DFX (Deferasirox), 0.05 mg/ml FAC, biological replicates of FAC and DFO treatment). (B) siRNA-mediated HERC2 knockdown attenuates NCOA4 degradation in response to iron loading. U2OS cells were treated with control or HERC2-specific RNAi for 72 hr, followed by 0.05 mg/ml FAC addition for the times indicated. NCOA4 protein level was evaluated by immunoblot. (C) U2OS cells were treated with 100 μg/ml (CHX) as indicated. To determine whether proteasome inhibition rescues NCOA4 turnover, cells were pre-treated with 1 μM Bortezomib (Btz) for 1 hr, followed by the addition of CHX for 60 min. NCOA4 stability was examined by immunoblot. (D) U2OS cells were pre-treated with DMSO (vehicle) or Bafilomycin A for 1 hr, followed by the addition of 100 μg/ml CHX for the time indicated. NCOA4 stability was evaluated by immunoblot.
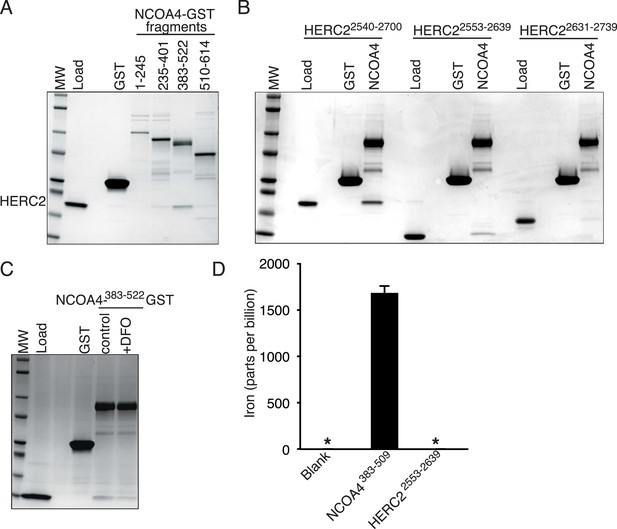
NCOA4 is an iron-binding protein.
(A) GST pulldown assay of recombinant NCOA4-GST fragments as indicated mixed with recombinant HERC22540−2700. Bound HERC2 was analyzed by 4–20% SDS-PAGE and Coomassie blue staining. Load is 5% of HERC2 input. (B) GST pulldown assay of recombinant NCOA4383−522-GST mixed with recombinant HERC2 fragments as indicated. Bound HERC2 was analyzed by 4–20% SDS-PAGE and Coomassie blue staining. Load is 5% of HERC2 input. (C) GST pulldown assay of recombinant NCOA4383−522-GST pre-treated as indicated with DFO and mixed with recombinant HERC2 CUL7 domain (amino acids 2553–2639). Bound HERC2 was analyzed by 4–20% SDS-PAGE and Coomassie blue staining. Load is 5% of HERC2 input. (D) NCOA4 NCOA4383−522-GST or the HERC22553−2639 (CUL7 domain) was expressed in Escherichia coli and the amount of co-purifying iron was measured by means of Inductively coupled plasma mass spectrometry (ICP-MS).
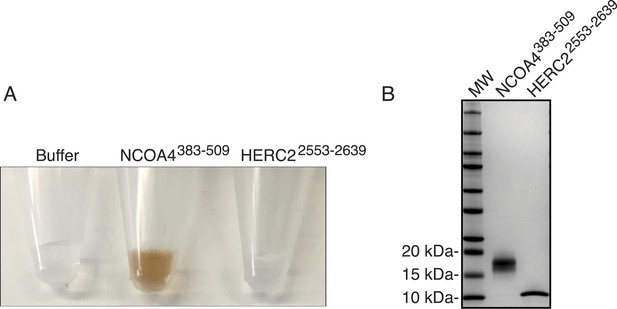
Evaluating NCOA4 iron binding.
(A) NCOA4383−509 and HERC22553−2639 were expressed in E. coli and the purified protein solution is depicted with NCOA4383−509 solution having a brown color suggestive of an iron-containing protein. (B) Coomassie stained SDS-PAGE gel of purified NCOA4383−509 and HERC22553−2639.
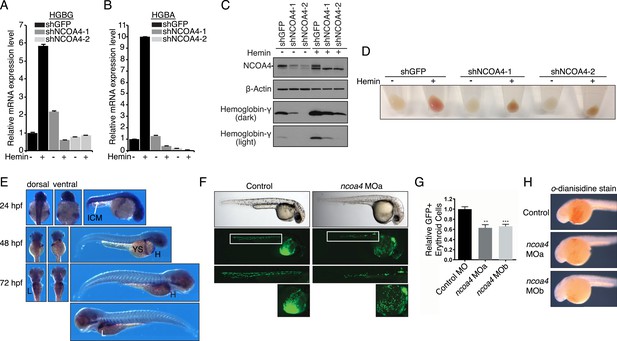
Knockdown of ncoa4 impacts erythropoiesis in zebrafish and tissue culture cells.
(A, B) Expression of HGBG (A), HGBA (B) determined by quantitative RT-PCR in K562 cells expressing a control (shGFP) or two independent shRNAs targeting NCOA4. Hemin treatment (25 μM, 72 hr) as indicated. (C) Immunoblot analysis of K562 cells as in (A) and (B) with indicated antibodies. Hemin treatment (25 μM, 72 hr) as indicated. (D) Appearance of K562 pellets as in (A, B, and C). Red coloration indicates appropriate hemoglobinization of cells after hemin differentiation. Brown coloration indicates accumulation of iron without hemoglobinization. (E) Expression of ncoa4 in circulating erythroid cells relative to sites of primitive hematopoiesis. Abbreviations: hpf = hours post-fertilization, ICM = intermediate cell mass, YS = yolk sac, H = heart, L = liver. (F) Circulating RBCs are visualized in globin-LCR:eGFP erythrocyte reporter zebrafish at 30 hpf. Morpholino-mediated knockdown of ncoa4 severely disrupts erythropoiesis (n > 30 each condition). Inset shows erythrocytes circulating in the caudal artery and caudal vein plexus. (G) FACS quantification of erythrocytes in globin-LCR:eGFP reporter zebrafish at 30 hpf following control or ncoa4-morpholino-mediated knockdown (ncoa4 MOa and MOb). ***p < 0.001 and **p < 0.002 by two-tailed unpaired t-test. (H) o-dianisidine staining (brown) for hemoglobinized red cells in zebrafish embryos. Embryos were grown from zygotes injected at the one- to two-cell stage with ncoa4-targeting morpholinos (MOa and MOb) or control injected zygotes (control). Ncoa4 MOa: 15/16 embryos with diminished staining in comparison to control. Ncoa4 MOb: 18/22 embryos with diminished staining in comparison to control.
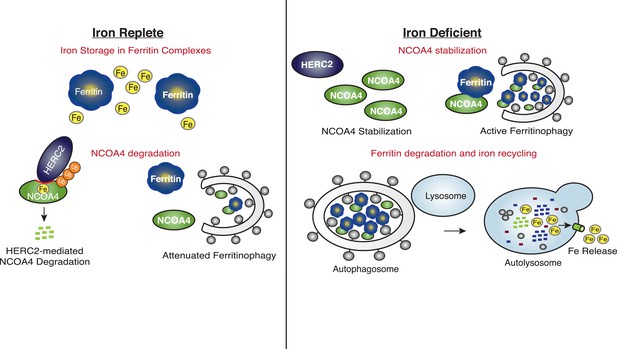
Iron Levels Regulate NCOA4-mediated ferritinophagy.
A model of NCOA4 ferritinophagy regulation (see the text for details).





