Evolutionary unique N-glycan-dependent protein quality control system plays pivotal roles in cellular fitness and extracellular vesicle transport in Cryptococcus neoformans
Figures
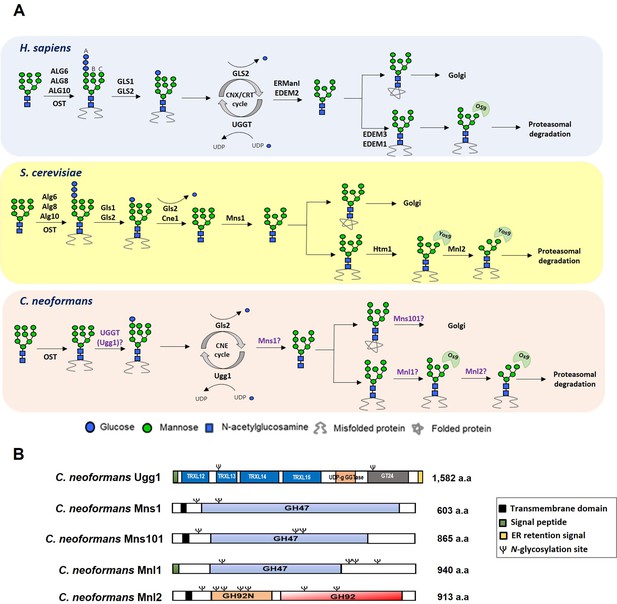
Presence of UDP-glucose:glycoprotein glucosyltransferase (UGGT) and α1,2-mannosidases as endoplasmic reticulum protein quality control (ERQC) components in Cryptococcus neoformans.
(A) Schematic representation of the ERQC pathway in Homo sapiens, Saccharomyces cerevisiae, and Cryptococcus neoformans. In mammals, the oligosaccharyltransferase (OST) complex attaches Glc3Man9GlcNAc2 to nascent polypeptides, followed by glucose trimming by glucosidases I and II (GLS1/GLS2). This generates Glc1Man9GlcNAc2, which binds to calnexin (CNX) and calreticulin (CRT) for folding. UGGT reglucosylates misfolded proteins, allowing refolding, while properly folded proteins undergo mannose trimming by ERManI before Golgi transport. In contrast, fungal ERQC systems differ in key components. S. cerevisiae lacks UGGT, relying instead on Gls1/Gls2 and calnexin (Cne1). C. neoformans possesses UGGT but lacks ER glucosyltransferases (Alg6, Alg8, and Alg19) and CRT, resulting in a distinct ERQC system. (B) Domain structures of proteins encoded by C. neoformans UGG1 (CNAG_03648), MNS1 (CNAG_02081), MNS101 (CNAG_03240), MNL1 (CNAG_01987), and MNL2 (CNAG_04498).
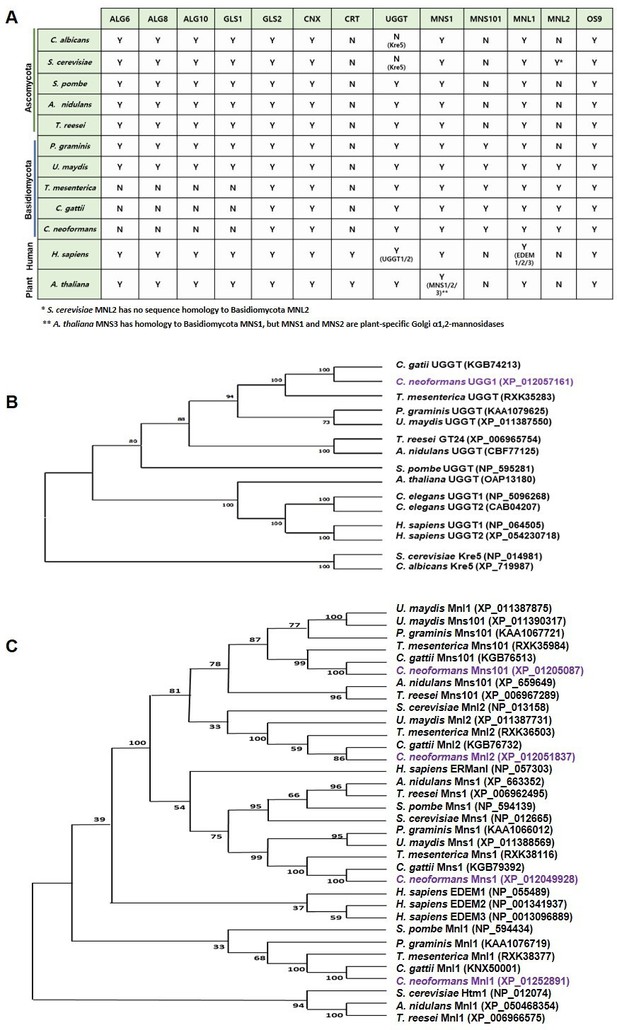
Evolutionary conservation and divergence of endoplasmic reticulum quality control (ERQC)-related gene homologs in eukaryotes.
(A) Distribution of ERQC homologs across various eukaryotic organisms. (B, C) Phylogenetic analysis of UDP-glucose:glycoprotein glucosyltransferase (UGGT), α1,2-mannosidase I (Mns1) and α1,2-mannosidase-like proteins (Mnl1/Htm1/Mnl2) homologs in different fungi. The phylogenetic trees were constructed using the maximum likelihood method in the MEGA X software with bootstrap analysis performed using 100 random resampling.

Domain analysis of putative C. neoformans endoplasmic reticulum quality control (ERQC)-associated proteins.
(A) Multiple sequence alignment of UGGT homologs generated using Clustal W 1.81. Identical and conserved amino acids are indicated by black and gray shading, respectively. The calcium-binding site and HDEL-like ER retention signal are highlighted with boxes. (B) Multiple sequence alignment highlighting the conserved cysteine and alanine residues (indicated by the arrows) present in all putative Mns1 and Mnl1 homologs, which are essential for mannosidase activity. This alignment was generated using Clustal W 1.81 and shaded using the GeneDoc software.
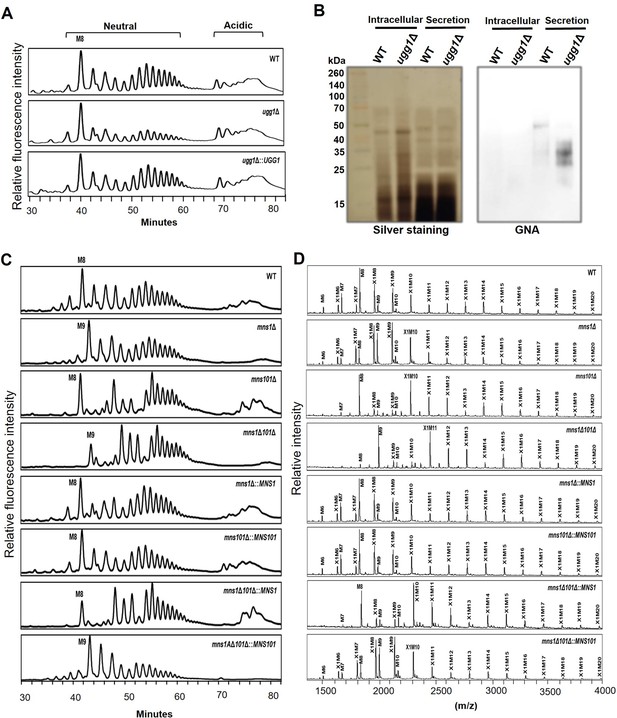
N-glycan profile analysis of C. neoformans endoplasmic reticulum quality control (ERQC) mutant strains.
(A) High-performance liquid chromatography (HPLC)-based analysis of N-glycan profiles of the ugg1Δ mutant. (B) Lectin blotting of sodium dodecyl sulfate (SDS)-polyacrylamide gels containing intracellular or proteins secreted into the culture supernatants of the wild type (WT) and ugg1Δ strains. Yeast cells were cultivated in YPD medium for 24 h, harvested, and subjected to sample preparation of soluble intracellular proteins and secreted proteins. The proteins (30 μg) were loaded on 15% SDS-polyacrylamide gel and analyzed using silver staining (left) or blotting (right) with Galanthus nivalis agglutinin conjugated to horseradish peroxidase (GNA-HRP, Roche). (C) HPLC analysis of total N-glycan profiles of mns1Δ, mns101Δ, and mns1Δ101Δ mutants. (D) Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) profiles of neutral N-glycans of mns1Δ, mns101Δ, and mns1Δ101Δ mutants. The N-glycans of cell wall mannoproteins from C. neoformans cells were AA-labeled and analyzed using HPLC. For MALDI-TOF analysis, neutral N-glycan fractions were obtained from the HPLC fractionation of total N-glycans.
-
Figure 2—source data 1
Original gel displayed in Figure 2B, indicating the relevant strains.
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig2-data1-v1.pdf
-
Figure 2—source data 2
Uncropped gel displayed in Figure 2B.
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig2-data2-v1.jpg
-
Figure 2—source data 3
Original blot membrane displayed in Figure 2B, indicating the relevant strains.
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig2-data3-v1.pdf
-
Figure 2—source data 4
Uncropped blot displayed in Figure 2B.
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig2-data4-v1.jpg
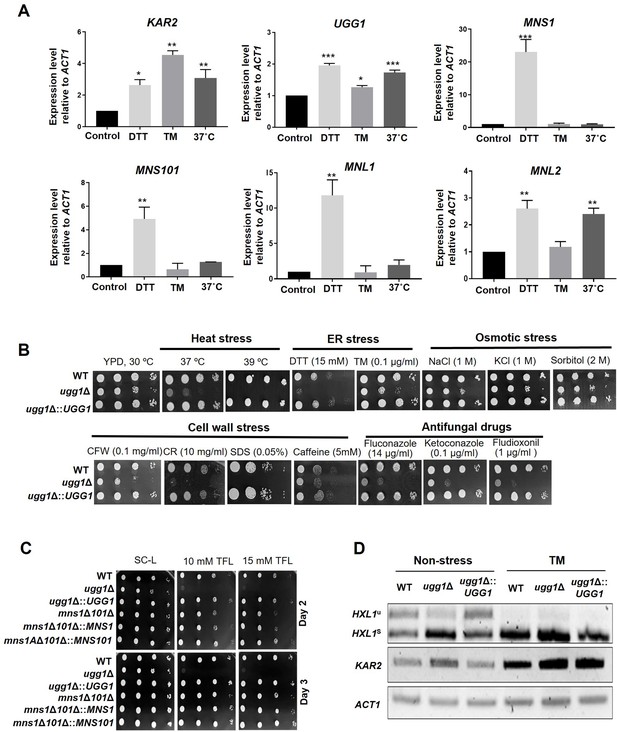
Growth phenotype of C. neoformans endoplasmic reticulum quality control (ERQC) mutant strains.
(A) Expression analysis of ERQC genes in C. neoformans. Yeast cells were cultured in YPD medium to a mid-logarithmic phase and exposed to dithiothreitol (DTT; 20 mM), tunicamycin (TM; 5 µg/ml), or cultured at 37 °C for 1 h. The relative transcript levels of C. neoformans genes were analyzed using qRT-PCR and normalized with that of ACT1. Error bars represent standard deviation of duplicated assays. All statistical data were determined based on one-way ANOVA and Dunnett’s post hoc test. ***p<0.0005, **p<0.003, ***p<0.005, *p<0.05. (B) Spotting analysis of C. neoformans ugg1Δ mutant strains under various stress conditions such as heat stress (37 °C and 39 °C), ER stress (DTT and TM), cell-wall stress (CFW: calcofluor white; CR: Congo red; SDS: sodium dodecyl sulfate; caffeine), osmotic stress (NaCl, KCl, sorbitol), and treatment with antifungal drugs (fluconazole, ketoconazole, fludioxonil). (C) Growth analysis in the presence of 5’,5’,5’-trifluoroleucine (TFL). Respective strains were spotted on SC-Leucine media with or without TFL supplementation. Plates were incubated for 3 days at 30 °C. (D) RT-PCR analysis of IRE1-dependent splicing of HXL1. Strains were cultured in YPD supplemented with 5 µg/ml TM.
-
Figure 3—source data 1
Raw data related to Figure 3.
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig3-data1-v1.zip
-
Figure 3—source data 2
Original gel displayed in Figure 3D (upper panel), indicating the relevant strains.
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig3-data2-v1.pdf
-
Figure 3—source data 3
Uncropped gels displayed in Figure 3D (upper panel).
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig3-data3-v1.jpg
-
Figure 3—source data 4
Original gel displayed in Figure 3D (middle panel), indicating the relevant strains.
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig3-data4-v1.pdf
-
Figure 3—source data 5
Uncropped gel displayed in Figure 3D (middle panel).
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig3-data5-v1.jpg
-
Figure 3—source data 6
Original gel displayed in Figure 3D (lower panel), indicating the relevant strains.
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig3-data6-v1.pdf
-
Figure 3—source data 7
Uncropped gel displayed in Figure 3D (lower panel).
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig3-data7-v1.jpg
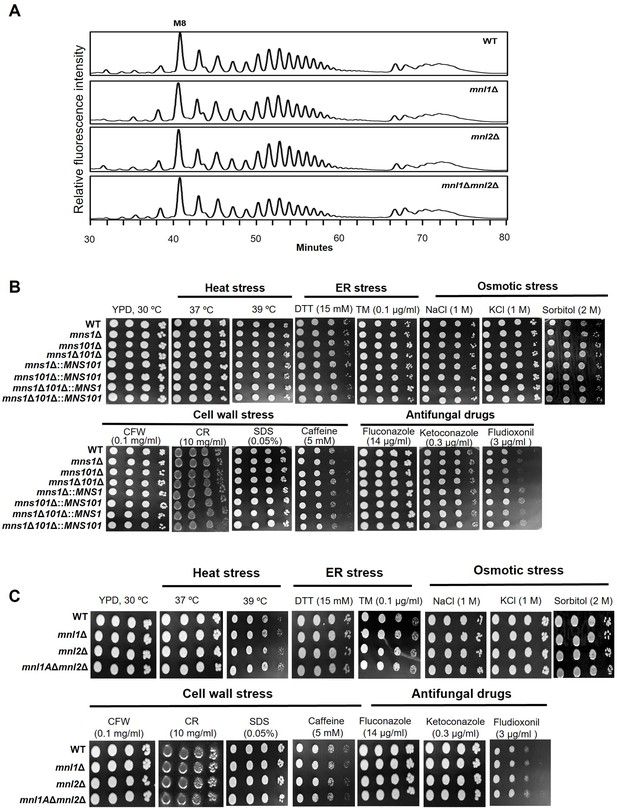
Phenotype characterization of C. neoformans α1,2-mannosidases.
(A) High-performance liquid chromatography (HPLC)-based N-glycan profiles of C. neoformans mnl1Δ, mnl2Δ, and mnl1Δmnl2Δ mutants. (B) Spotting analysis of C. neoformans mns1Δ, mns101Δ, and mns1Δ101Δ mutants cultivated under various stress conditions including heat stress (37 °C, 39 °C), ER stress (DTT: dithiothreitol; TM: tunicamycin), cell-wall stress (CFW: calcofluor white; CR: Congo red; SDS: sodium dodecyl sulfate, caffeine), osmotic stress (NaCl, KCl, sorbitol), and treatment with antifungal drugs (fluconazole, ketoconazole, fludioxonil). (C) Spotting analysis of C. neoformans mnl1Δ, mnl2Δ, and mnl1Δmnl2Δ cultivated under various stress-inducing conditions.
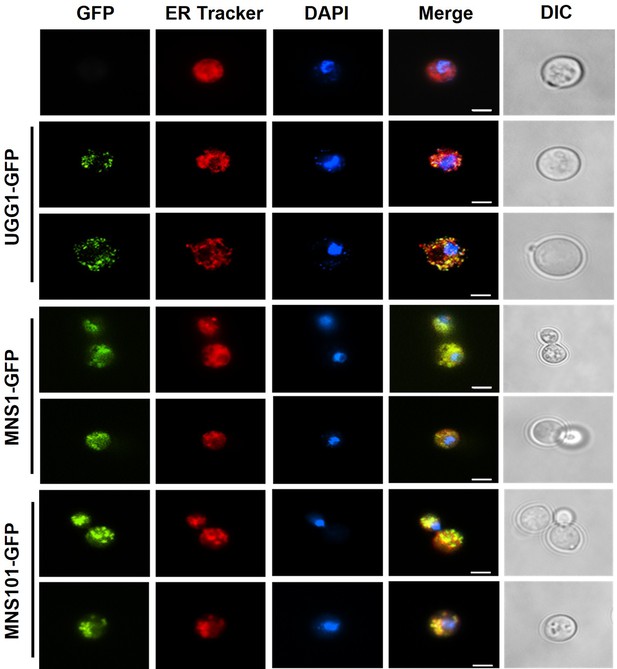
Subcellular localization of GFP-tagged Ugg1, Mns1, and Mns101.
ER tracker was used to visualize the ER membrane. Scale bar, 2.5 μm.
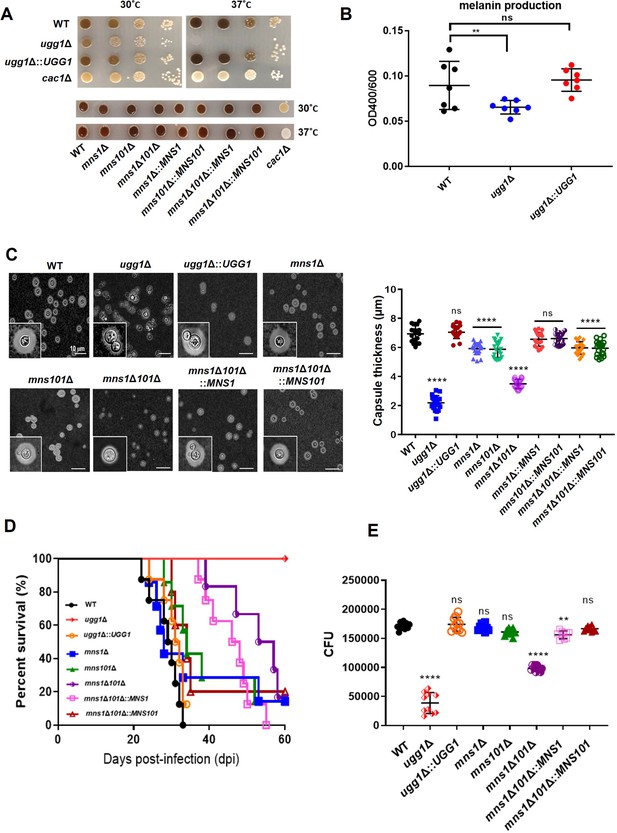
In vitro and in vivo virulence-associated phenotypes of C. neoformans UGG1, MNS1, and MNS101 mutant strains.
(A) Melanin synthesis analysis on l-DOPA plates. WT, ugg1Δ, ugg1Δ::UGG1, mns1Δ, mns101Δ, mns1Δ101Δ, mns1Δ101Δ::MNS1, mns1Δ101Δ::MNS101, and cac1Δ (negative control) strains were serially diluted, plated on l-DOPA plates, and incubated at 30 °C and 37 °C. (B) Melanin synthesis activity per cell. WT, ugg1Δ, ugg1Δ::UGG1 were cultured in liquid l-DOPA medium. The amount of melanin in the culture supernatant was measured and normalized by cell density. (C) Capsule formation. Cells were cultured for 2 days in 10% Sabouraud media at 30 °C and observed under the microscope. Statistical significance: ****p<0.0001, ns, not significant. (D) In vivo virulence analysis. A/Jcr mice (n = 8) were infected with 105 cells of WT, ugg1Δ, and ugg1Δ::UGG1, mns1Δ, mns101Δ, mns1Δ101Δ, mns1Δ101Δ::MNS1, and mns1Δ101Δ::MNS101 strains, and survival was monitored for 2 months. (E) Survival of C. neoformans in macrophages. Survival of C. neoformans cells within the J774A.1 macrophage-like cell line was determined by counting colony-forming unit (CFU) obtained from lysed macrophages from two biologically independent experiment sets. ****p<0.0001, ***p<0.0005, *p<0.05, ns, not significant. All statistical data were determined based on one-way ANOVA and Dunnett’s post hoc test.
-
Figure 4—source data 1
Raw data related to Figure 4.
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig4-data1-v1.zip
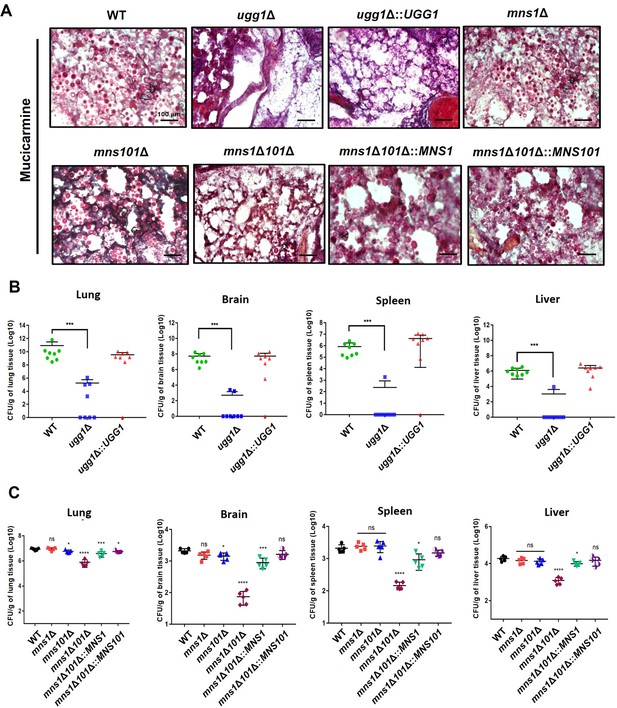
Fungal burden of mice infected with C. neoformans endoplasmic reticulum quality control (ERQC)-related mutant cells.
(A) Histopathological analysis of lung tissue infected with C. neoformans. Lungs were excised at the death point (60 dpi) and stained with mucicarmine to visualize C. neoformans cells. (B) Distribution of C. neoformans cells (WT, ugg1Δ, and ugg1Δ::UGG1) in systemic organs at the humane endpoint. The fungal burden was determined using colony-forming unit (CFU) counts after plating on YPD medium supplemented with chloramphenicol. (C) Fungal burden assay of WT, mns1Δ, mns101Δ, and mns1Δ101Δ at the early stage of infection (7 dpi). Statistical significance: ****p<0.0001, ***p<0.0005, *p<0.05, ns, not significant. All statistical data were determined by one-way ANOVA and Dunnett’s post hoc test.
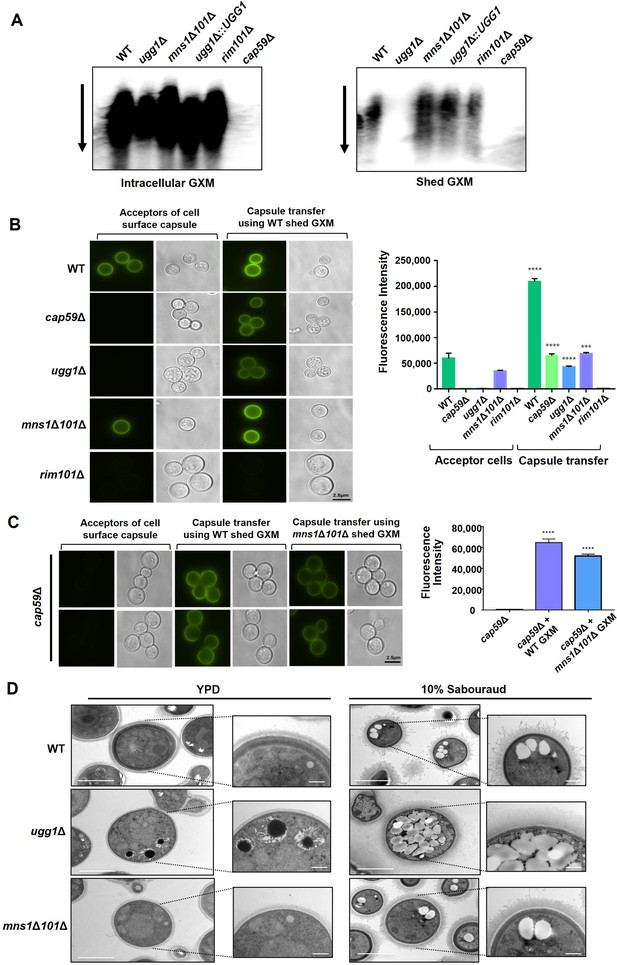
Capsule shedding and transfer analysis of C. neoformans UGG1, MNS1, and MNS101 mutant strains.
(A) Capsule shedding analysis. The presence of intracellular (left) and shed (right) glucuronoxylomannan (GXM) was assessed by blotting a cell culture filtrate using the monoclonal antibody 18B7. The arrow indicates the direction of electrophoresis. (B) Capsule transfer analysis using exogenous capsule material from WT. The capsule transfer assay was performed using the indicated strains as acceptors. Surface capsules were probed using the anti-GXM antibody 18B7 conjugated with AlexaFluor 488. Quantitative measurements of fluorescence intensity were calculated based on independent triplicate experiments with standard deviations presented as error bars. (C) Capsule transfer analysis using exogenous capsule material from WT or mns1Δ101Δ. Statistical significance: ****p<0.0001. All statistical data were determined based on one-way ANOVA and Dunnett’s post hoc test. (D) Transmission electron microscopy (TEM) of C. neoformans WT, ugg1Δ, and mns1Δ101Δ strains. Yeast cells were grown overnight at 30 °C in YPD medium and fixed in 2% glutaraldehyde and 2% paraformaldehyde. A Zeiss Axioscope (A1) equipped with an AxioCan MRm digital camera was used to visualize India ink-stained C. neoformans cells. Specimens were prepared using critical point drying prior to TEM imaging. Capsule and yeast cell body diameters were measured using ImageJ (National Institute of Health).
-
Figure 5—source data 1
Raw data related to Figure 5.
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig5-data1-v1.zip
-
Figure 5—source data 2
Uncropped membrane blot displayed in Figure 5A (left panel), indicating the relevant strains.
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig5-data2-v1.pdf
-
Figure 5—source data 3
Uncropped membrane blot displayed in Figure 5A (left panel).
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig5-data3-v1.jpg
-
Figure 5—source data 4
Uncropped membrane blot displayed in Figure 5A (right panel), indicating the relevant strains.
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig5-data4-v1.pdf
-
Figure 5—source data 5
Uncropped membrane blot displayed in Figure 5A (right panel).
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig5-data5-v1.jpg
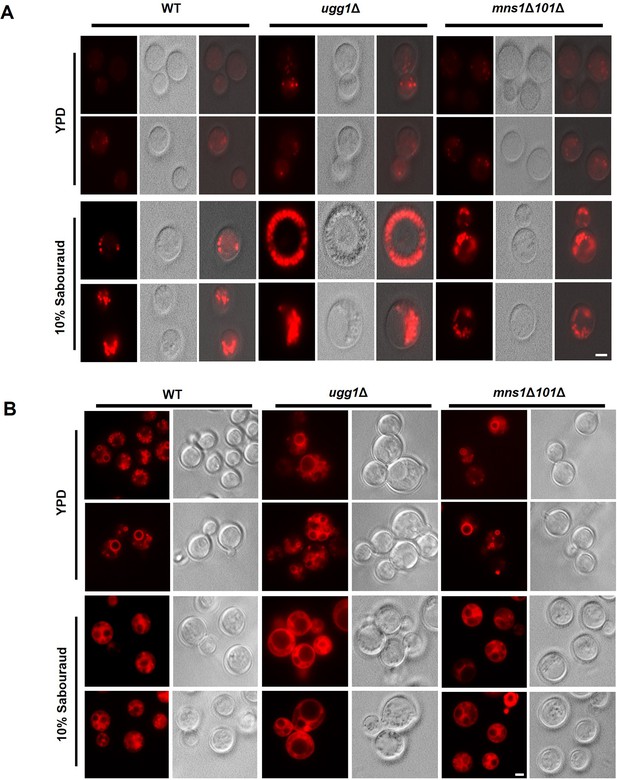
Lipid droplets and vacuole staining of WT and ugg1Δ strains.
Respective strains were cultivated in either YPD or 10% Sabouraud media at 30 °C and washed twice with PBS. Cells (OD600=1) were stained with LipidTox (Invitrogen, USA) for detection of LDs (A) or FM4-64 (Invitrogen) for detection of vacuoles (B), for 30 min at room temperature in the dark. Cells were visualized using an Eclipse Ti-E fluorescence microscope (Nikon, Japan), equipped with a Nikon DS-Qi2 camera and a Plan Apo VC ×100 oil differential interference contrast (DIC) lens. Images were processed using the NIS-Elements imaging software (Nikon). All images are shown on the same scale. Scale bar, 2.5 µm.
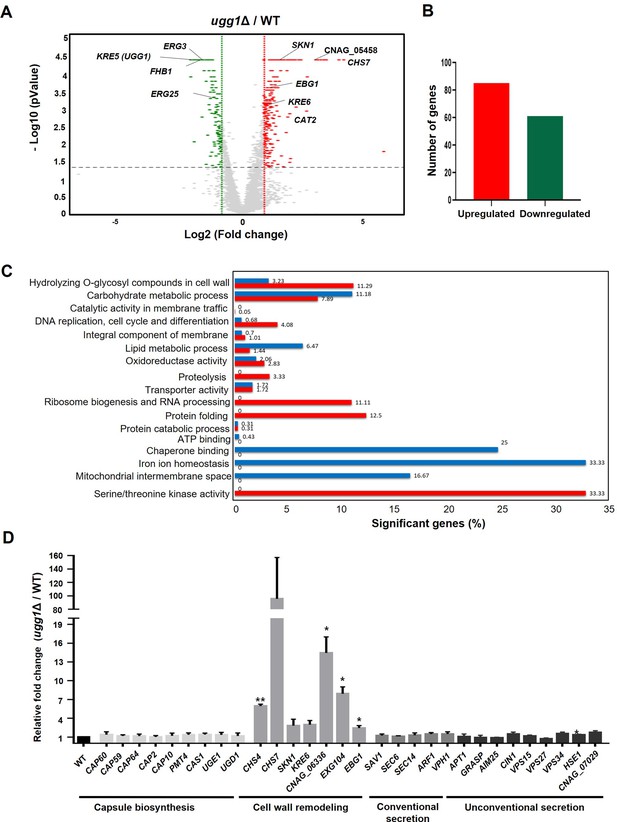
Transcriptome analysis of C. neoformans WT and ugg1Δ cells.
(A) Volcano plot comparing a twofold differential gene expression between ugg1Δ and WT strains under normal growth conditions. (B) Number of genes upregulated and downregulated by ≥2-fold in ugg1Δ compared with that of the WT. (C) Gene ontology (GO) analysis of differentially expressed genes between WT and ugg1Δ strains. Significantly upregulated genes in ugg1Δ are shown in red, whereas significantly downregulated genes in ugg1Δ are shown in blue. (D) qRT-PCR analysis of mRNA expression levels of a set of genes responsible for capsule biosynthesis, cell wall remodeling, and both conventional and non-conventional secretion in ugg1Δ vs WT under normal growth conditions. Error bars represent standard deviation from three biologically independent experiment sets.
-
Figure 6—source data 1
Raw data related to Figure 6.
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig6-data1-v1.zip
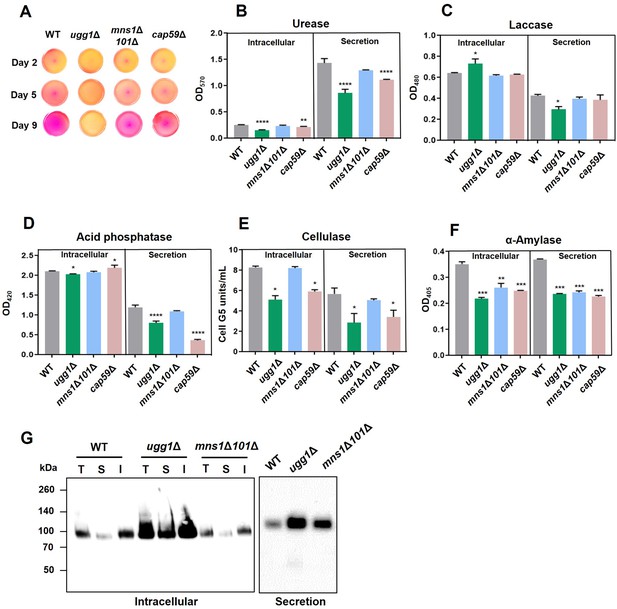
Analysis of protein secretion in C. neoformans UGG1, MNS1, and MNS101 mutant strains.
(A) Spot assay for urease analysis on Christensen’s urea agar. Absence of pink coloration indicates loss of urease activity. (B–D) Analysis of secretion of the virulence-related enzymes urease, laccase, and acid phosphatase from three biologically independent experiment sets. Statistical significance: ****p<0.0001, **p<0.003, ***p<0.005, *p<0.05. All statistical data were determined based on one-way ANOVA and Dunnett’s post hoc test. (E, F) Analysis of secretion for non-virulence-related enzymes such as cellulase and α-amylase. Statistical significance: ***p<0.0005, **p<0.003, *p<0.05, ns, not significant. All statistical data were determined based on one-way ANOVA and Dunnett’s post hoc test. (G) Analysis of the conventional secretion of Cda1 in C. neoformans. Presence of Cda1 was analyzed in total (T), soluble (S), and insoluble (I) fractions of intracellular extracts (left), along with the secreted fraction (right). Subcellular fractionations were performed as previously described (Thak et al., 2022), and the fractions were subjected to western blotting analysis using an anti-Cda1 antibody.
-
Figure 7—source data 1
Raw data related to Figure 7.
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig7-data1-v1.zip
-
Figure 7—source data 2
Uncropped membrane blot displayed in Figure 7G (left panel), indicating the relevant strains.
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig7-data2-v1.pdf
-
Figure 7—source data 3
Uncropped membrane blot displayed in Figure 7G (left panel).
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig7-data3-v1.jpg
-
Figure 7—source data 4
Uncropped membrane blot displayed in Figure 7G (right panel), indicating the relevant strains.
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig7-data4-v1.pdf
-
Figure 7—source data 5
Uncropped membrane blot displayed in Figure 7G (right panel).
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig7-data5-v1.jpg
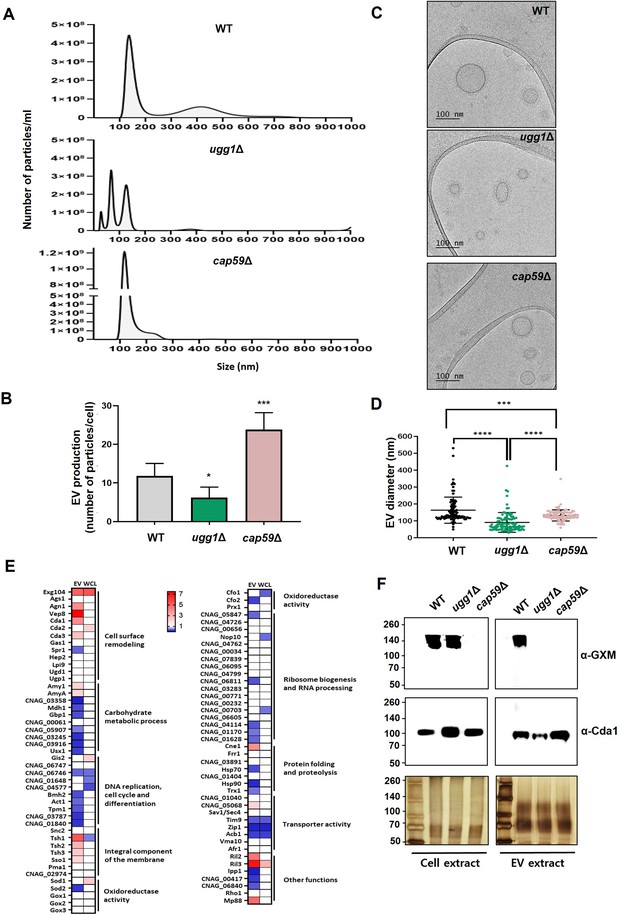
Analysis of extracellular vesicles (EVs) purified from WT, ugg1Δ, and cap59Δ cells.
(A, B) Nanoparticle tracking analysis (NTA) of EVs extracted from WT, ugg1Δ, and cap59Δ strains and quantification of total EV concentration per cell density. Quantitative measurements were derived from three independent experiments with standard deviations presented as error bars. Statistical significance: ***p<0.0005, *p<0.05. All statistical data were determined based on one-way ANOVA and Dunnett’s post hoc test. (C, D) Cryo-TEM imaging of purified EVs and comparative analysis of EV size in WT, ugg1Δ, and cap59Δ strains. Scale bar, 100 nm. The outer EV diameter of a total number of 100 EVs per strain, captured using cryo-TEM, were measured. (E) Heatmap representation of fold change between WT and ugg1Δ EV-associated proteins, commonly detected in this study and in previously reported EV proteome datasets (ugg1Δ/WT). Upregulated proteins in ugg1Δ are shown in red, whereas downregulated proteins are shown in blue. The proteome data of whole-cell lysates (WCL), generated from the cell pellets obtained after EV separation, were included for comparison. (F) Blotting analysis of GXM in the C. neoformans cells and EVs of WT, ugg1Δ, and cap59Δ strains. The 8 M urea extracts were obtained from EVs and cell pellets, from which EVs are generated. The urea extracts (5 μg total proteins) were loaded on 8% SDS-polyacrylamide gel and subjected to silver staining or blotting analysis using the anti-GXM 18B7 (α-GXM) and anti-Cda1 (α-Cda1) antibodies, respectively. Left: total cell extract. Right: total EV extract.
-
Figure 8—source data 1
List of proteins identified in the proteomic analysis of the extracellular vesicles (EVs) and whole-cell lysates (WCL) obtained from wild type (WT) and ugg1Δ.
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig8-data1-v1.xlsx
-
Figure 8—source data 2
List of proteins identified in the proteomic analysis of the wild type (WT) and ugg1Δ secretome and whole-cell lysates (WCL).
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig8-data2-v1.xlsx
-
Figure 8—source data 3
Original gel displayed in Figure 8F (left panel), indicating the relevant strains.
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig8-data3-v1.pdf
-
Figure 8—source data 4
Uncropped gels displayed in Figure 8F (left panel).
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig8-data4-v1.jpg
-
Figure 8—source data 5
Original gel displayed in Figure 8F (right panel), indicating the relevant strains.
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig8-data5-v1.pdf
-
Figure 8—source data 6
Uncropped gels displayed in Figure 8F (right panel).
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig8-data6-v1.jpg
-
Figure 8—source data 7
Original gel displayed in Figure 8F (left, upper panel), indicating the relevant strains.
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig8-data7-v1.pdf
-
Figure 8—source data 8
Uncropped gels displayed in Figure 8F (left, upper panel).
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig8-data8-v1.jpg
-
Figure 8—source data 9
Original gel displayed in Figure 8F (left, middle panel), indicating the relevant strains.
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig8-data9-v1.pdf
-
Figure 8—source data 10
Uncropped gels displayed in Figure 8F (left, middle panel).
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig8-data10-v1.jpg
-
Figure 8—source data 11
Original gel displayed in Figure 8F (right, upper panel), indicating the relevant strains.
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig8-data11-v1.pdf
-
Figure 8—source data 12
Uncropped gels displayed in Figure 8F (right, upper panel).
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig8-data12-v1.jpg
-
Figure 8—source data 13
Original gel displayed in Figure 8F (right, middle panel), indicating the relevant strains.
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig8-data13-v1.pdf
-
Figure 8—source data 14
Uncropped gels displayed in Figure 8F (right, middle panel).
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig8-data14-v1.jpg
-
Figure 8—source data 15
Raw data related to Figure 8.
- https://cdn.elifesciences.org/articles/103729/elife-103729-fig8-data15-v1.zip
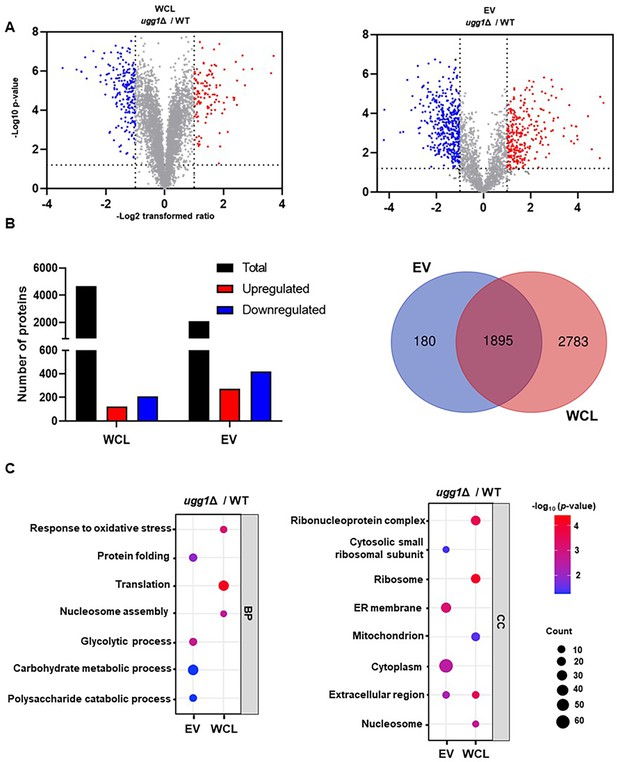
Comparative proteomic analysis of whole-cell lysate (WCL) and extracellular vesicles (EVs) from WT and ugg1Δ strains.
EVs were isolated from C. neoformans cells cultivated on synthetic dextrose (SD) solid medium for 24 h. (A) Volcano plot highlighting proteins exhibiting a >2-fold change in expression. (B) Total number of proteins identified in WCL and EV samples, with red and blue columns representing proteins with >2-fold differential expression. (C) Gene ontology (GO) enrichment analysis of differentially expressed proteins (>2-fold), categorized into biological process (BP) and cellular component (CC). Bubble plots were generated using the SRPlot web tool (http://www.bioinformatics.com.cn/srplot; Tang et al., 2023).
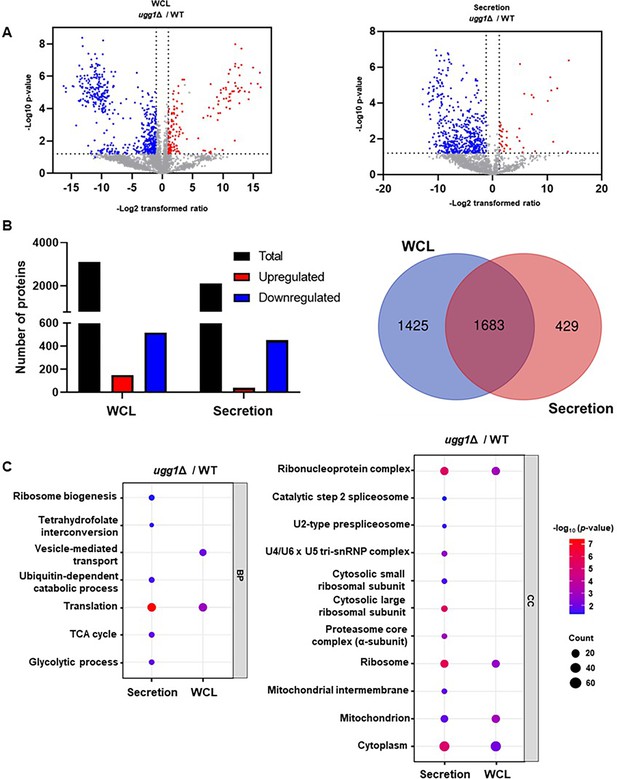
Comparative proteomic analysis of whole-cell lysate (WCL) and secretion fractions from WT and ugg1Δ strains.
C. neoformans cells were cultivated in synthetic dextrose (SD) broth for 24 h. Secretome samples were prepared by TCA precipitation of culture supernatants. (A) Volcano plot highlighting proteins exhibiting a >2-fold change in expression. (B) Total number of proteins identified in WCL and extracellular vesicle (EV) samples, with red and blue columns representing proteins with >2-fold differential expression. (C) Gene ontology (GO) enrichment analysis of differentially expressed proteins (>2-fold), categorized into biological process (BP) and cellular component (CC). Bubble plots were 60 generated using the SRPlot web tool (http://www.bioinformatics.com.cn/srplot; Tang et al., 2023).
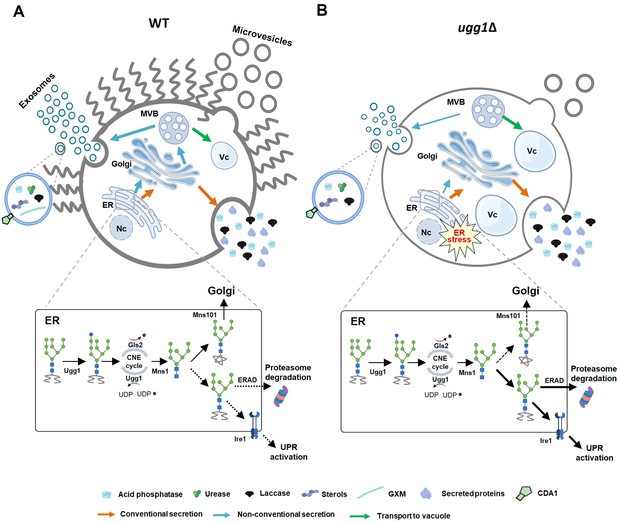
Impact of endoplasmic reticulum quality control (ERQC) disruption on glycoprotein folding and extracellular vesicle (EV)-mediated transport of virulence factors in C. neoformans.
(A) In WT strain, C. neoformans UGGT homolog, Ugg1, functions as a sensor for misfolded glycoproteins within the ER, playing a crucial role in protein quality control. Functional ERQC is essential not only for ensuring the proper folding of glycoproteins, which is critical for maintaining cellular fitness, but also for facilitating EV-mediated secretion of capsule polysaccharides and virulence-related enzymes necessary for pathogenicity. (B) In the UGGT-deficient strain (ugg1Δ), ER stress is increased because of misfolded protein accumulation within the ER lumen. This heightened stress leads to decreased cellular fitness, which negatively impacts EV biogenesis and cargo loading. Consequently, significant defects occur in EV-mediated transport, which ultimately leads to a complete loss of virulence. Nc: nucleus; Vc: vacuoles. This figure was partially created using BioRender (https://BioRender.com/3gzzput, https://BioRender.com/bgibgte).
Additional files
-
Supplementary file 1
List of strains, plasmids and primers.
(A) List of strains used in this study. (B) List of plasmids used in this study. (C) List of primers used in this study.
- https://cdn.elifesciences.org/articles/103729/elife-103729-supp1-v1.docx
-
Supplementary file 2
List of Cryptococcus neoformans genes showing differential expression between ugg1Δ and wild type strains.
(A) Upregulated (>2-fold) Cryptococcus neoformans genes in ugg1Δ than that of wild type (WT) cultivated in yeast extract peptone dextrose (YPD) at 30 °C. (B) Downregulated (>2-fold) Cryptococcus neoformans genes in ugg1Δ than that of wild type (WT) cultivated in YPD at 30 °C.
- https://cdn.elifesciences.org/articles/103729/elife-103729-supp2-v1.docx
-
Supplementary file 3
Representative extracellular vesicle (EV)-associated proteins commonly detected in this study and previously reported Cryptococcus neoformans EV proteome data sets.
- https://cdn.elifesciences.org/articles/103729/elife-103729-supp3-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/103729/elife-103729-mdarchecklist1-v1.docx





