Chalkophore-mediated respiratory oxidase flexibility controls M. tuberculosis virulence
Figures
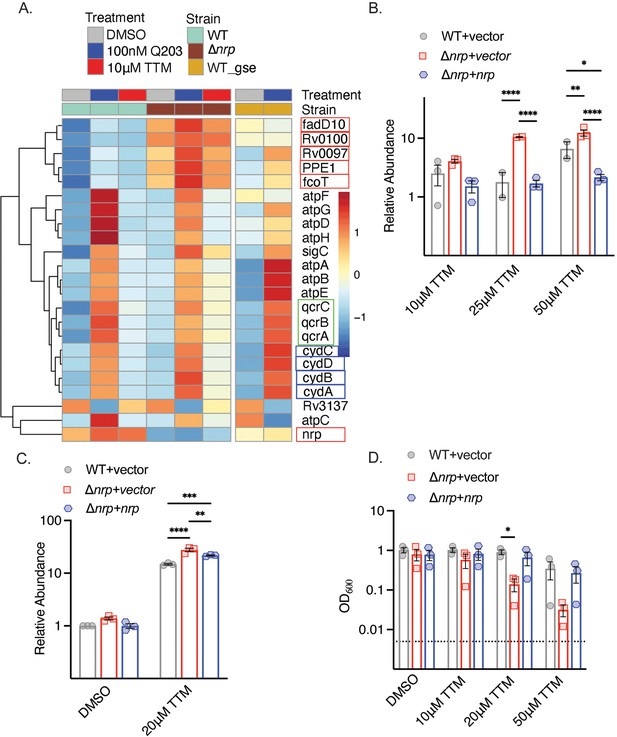
Copper deprivation in chalkophore-deficient M. tuberculosis mimics bcc:aa3 oxidase inhibition.
(A) Heat map of transcripts encoding selected respiratory chain components determined by RNA sequencing of M. tuberculosis wild-type (WT) or Δnrp treated with TTM or Q203. WT_GSE is the published dataset GSE159080 of M. tuberculosis H37Rv treated with Q203. Genes in the chalkophore cluster are boxed in red, genes encoding the cytochrome BD (CytBD) oxidase in blue, and genes encoding components of the bcc:aa3 supercomplex are in green. (B) RT-qPCR of the transcript encoding CydA in M. tuberculosis WT, Δnrp, and complemented strain treated with varying TTM concentrations for 4 hr. Error bars represent the standard error of the mean (SEM). Statistical significance determined via two-way ANOVA with Tukey correction for multiple comparisons. *p<0.05, **p<0.01, ****p<0.0001. (C) RT-qPCR of the transcript encoding CydA in M. tuberculosis WT, Δnrp, and complemented strain treated with 20 μM TTM for 24 hr. Error bars are SEM. Statistical significance determined via two-way ANOVA with Tukey correction for multiple comparisons. **p<0.01, ***p<0.001, ****p<0.0001. (D) Dose-dependent effect of tetrathiomolybdate (TTM) on growth of the indicated M. tuberculosis strains at 7 d post inoculation. The dotted line indicates the starting inoculum. Error bars are SEM. Statistical significance determined via two-way ANOVA with Tukey correction for multiple comparisons. *p<0.05.
-
Figure 1—source data 1
Raw data values for Figure 1B–D.
- https://cdn.elifesciences.org/articles/105794/elife-105794-fig1-data1-v1.xlsx
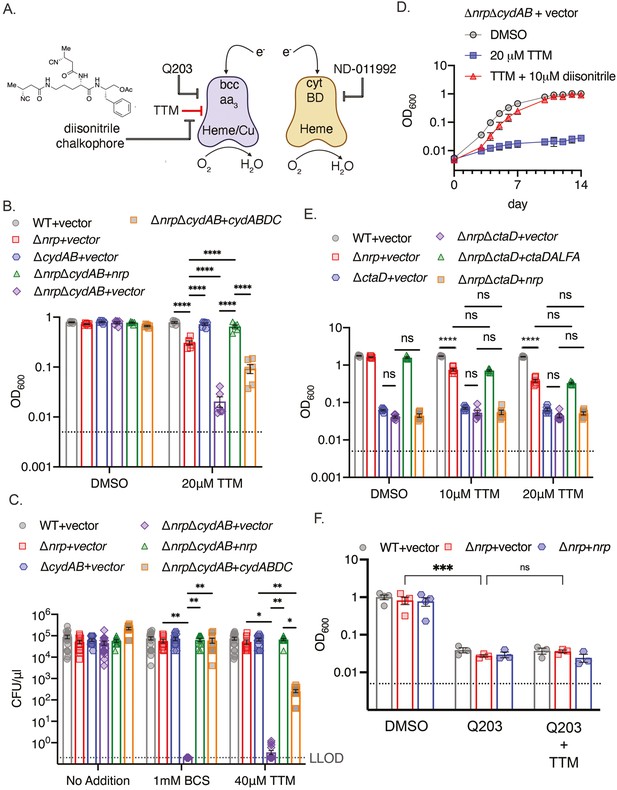
Chalkophores maintain M. tuberculosis viability through the heme-copper bcc:aa3 oxidase during copper starvation.
(A) Schematic of the terminal respiratory oxidases of M. tuberculosis. The bcc:aa3 oxidase is a heme-copper oxidase and cytochrome BD (CytBD) is a copper-independent heme oxidase. Both transfer electrons to oxygen. Q203 is an inhibitor of bcc:aa3 by targeting the QcrB subunit, whereas ND-011992 targets CytBD. The two oxidases are individually dispensable due to compensation by the other oxidase, but M. tuberculosis lacking both is nonviable. The model to be tested is that copper chelation deprives the bcc:aa3 oxidase of copper and that diisonitrile chalkophores counter this copper deprivation stress. (B) Liquid growth assays of the indicated strains with or without 20 µM TTM treatment. OD600 at day 10 post-inoculation displayed. Dotted line indicates starting inoculum. Error bars are SEM. Statistical significance determined via two-way ANOVA with Tukey correction for multiple comparisons. ****p<0.0001. (C) Bacterial survival of the indicated strains on agar media containing no addition, 1 mM BCS, or 40 μM TTM. Dotted line indicates lower limit of detection (LLOD). Error bars are SEM. Statistical significance determined via two-way ANOVA with Tukey correction for multiple comparisons. **p<0.01, *p<0.05 (D) The copper deprivation sensitivity of M. tuberculosis ΔnrpΔcydAB strain can be rescued with a synthetic diisonitrile chalkophore. Liquid growth assays of ΔnrpΔcydAB with DMSO, 20 μM TTM, or 20 μM TTM with 10 μM of the diisonitrile chalkophore pictured in panel A. Error bars are SEM. (E) The bcc:aa3 oxidase is the only target of copper starvation countered by diisonitrile chalkophores. Liquid growth assays of the indicated strains treated with 10 or 20 µM TTM, or DMSO vehicle control. OD600 at day 10 post-inoculation displayed. Dotted line indicates starting inoculum. Error bars are SEM. Statistical significance determined via two-way ANOVA with Tukey correction for multiple comparisons. ns = not significant, ****p<0.0001. (F) The effect of copper deprivation is masked by inhibition of QcrB subunit of bcc:aa3. Liquid growth assays of the indicated strains treated with Q203 (100 nM) alone or co-treated with 100 nM Q203 and 10 μM TTM. OD600 at day 7 post-inoculation displayed. Dotted line indicates starting inoculum. Error bars are SEM. Statistical significance determined via two-way ANOVA with Tukey correction for multiple comparisons. ***p<0.001, ns = not significant.
-
Figure 2—source data 1
Raw data for Figure 2B–F.
- https://cdn.elifesciences.org/articles/105794/elife-105794-fig2-data1-v1.xlsx
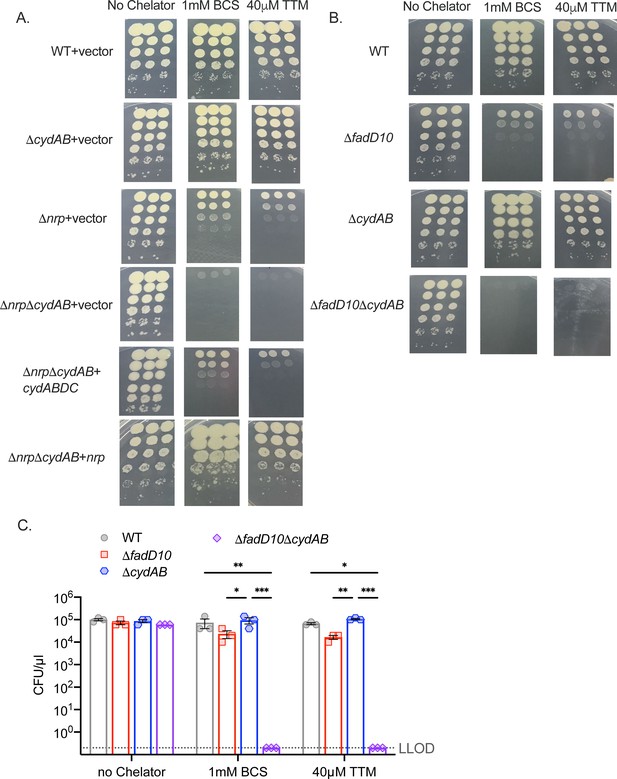
Loss of cydAB severely sensitizes chalkophore-deficient M. tuberculosis to copper chelation.
(A, B) Images of triplicate 10-fold serial dilutions of the indicated M. tuberculosis strains on agar media containing no chelator, 1 mM bathocuproinedisulfonic acid (BCS), or 40 µM tetrathiomolybdate (TTM). (C) Quantitation of the agar survival assay in panel B. Error bars are SEM. Statistical significance determined by two-way ANOVA with Tukey correction for multiple comparisons. *p<0.05, **p<0.01, ***p<0.001.
-
Figure 2—figure supplement 1—source data 1
Raw bacterial counts from the experiment in Figure 2—figure supplement 1C.
- https://cdn.elifesciences.org/articles/105794/elife-105794-fig2-figsupp1-data1-v1.xlsx
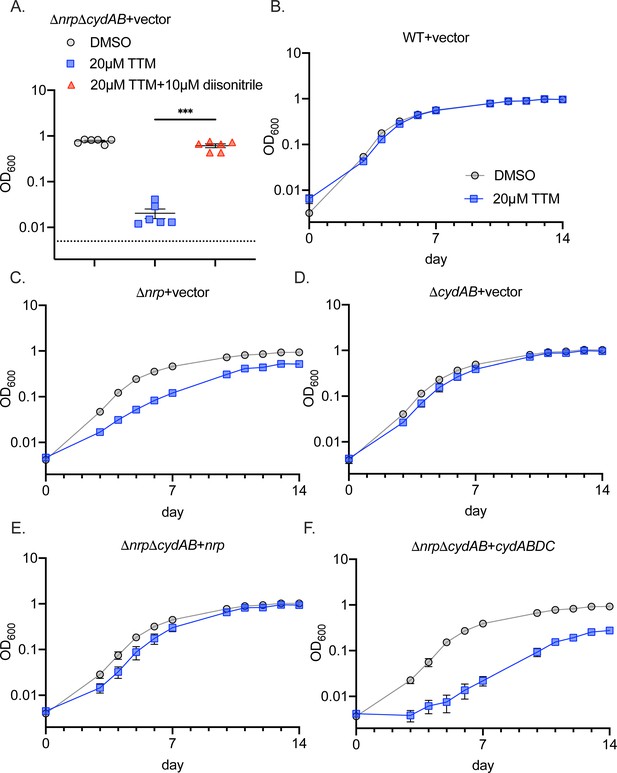
Full growth curves of chalkophore deficient strains with TTM copper chelation.
(A) Quantitation of optical density in liquid culture from Figure 2D at day 10 time point of the ΔnrpΔcydAB strain treated with DMSO, tetrathiomolybdate (TTM), or TTM + synthetic diisonitrile. Error bars are SEM. Statistical significance determined by two-way ANOVA with Tukey correction for multiple comparisons ***p<0.001. Dotted line indicates starting inoculum. (B–F) Full growth curves in liquid media quantitated in Figure 2B for the indicated strains treated with DMSO (gray symbols) or 20 µM TTM (blue symbols). Error bars are SEM and if not visible are within the graphed symbol.
-
Figure 2—figure supplement 2—source data 1
Raw data showing OD600 measuremenets graphed in Figure 2—figure supplement 2A–F.
- https://cdn.elifesciences.org/articles/105794/elife-105794-fig2-figsupp2-data1-v1.xlsx
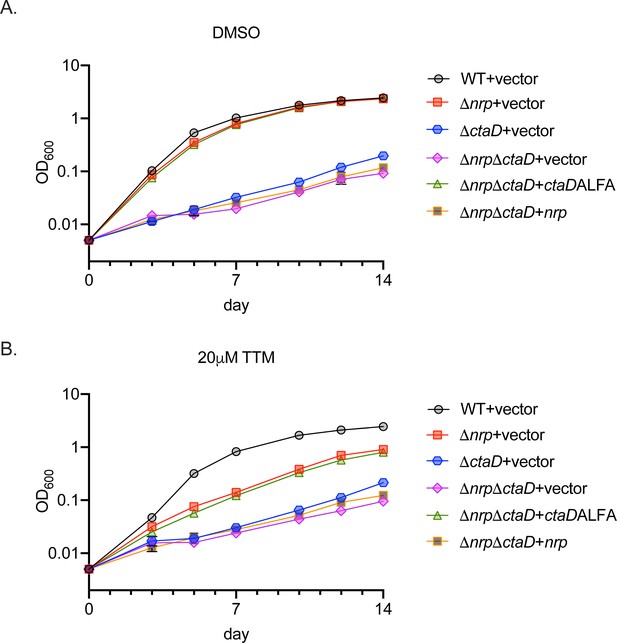
Full growth curves in basal and copper chelation conditions of ctaD mutant strains.
(A-B) Full growth curves in liquid media quantitated in Figure 2E for the indicated strains treated with DMSO (A) or 20 µM tetrathiomolybdate (TTM) (B). Error bars are SEM and are within the symbol if not visible.
-
Figure 2—figure supplement 3—source data 1
Raw data showring OD600 values for Figure 2—figure supplement 3A and B.
- https://cdn.elifesciences.org/articles/105794/elife-105794-fig2-figsupp3-data1-v1.xlsx
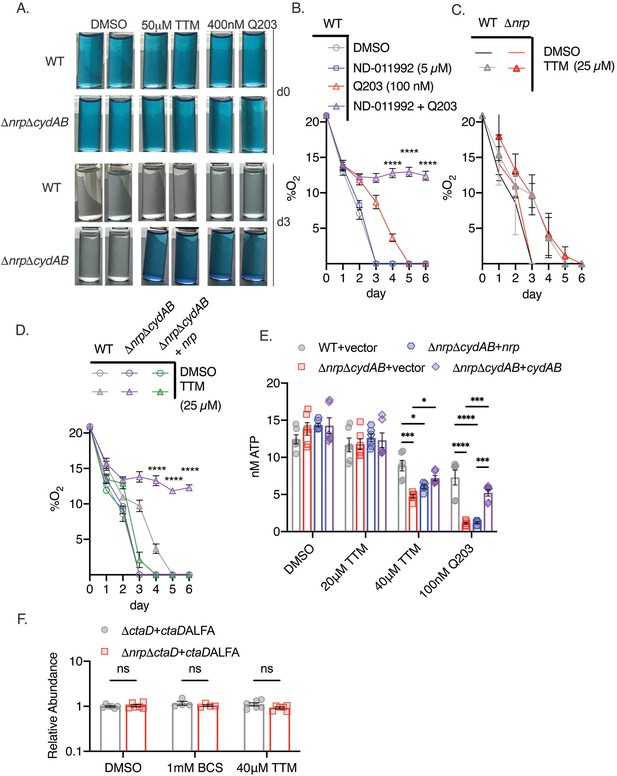
Chalkophore biosynthesis maintains oxidative phosphorylation through the heme-copper bcc:aa3 oxidase.
(A) Methylene blue decolorization assay of oxygen consumption under copper deprivation (tetrathiomolybdate, TTM) or treatment with Q203 in wild-type (WT) or ΔnrpΔcydAB M. tuberculosis at day 0 (d0) or day 3 (d3) of incubation. Clear vials indicate oxygen consumption by respiration. (B) Quantitative measurement of oxygen consumption using oxygen-sensitive optical sensors. WT M. tuberculosis treated with DMSO, ND-011992, Q203, or both ND-011992 and Q203. Oxygen measurements were taken daily. Each point represents three measurements of two biological replicates. Error bars are SEM. Statistical significance between Q203 and ND-011992 +Q203 determined via two-way ANOVA with Tukey correction for multiple comparisons. ****p<0.0001. (C) Same assay as in panel B with WT and Δnrp M. tuberculosis treated with DMSO or 25 μM TTM. Error bars are SEM. (D) Same assay as in panel B with WT, ΔnrpΔcydAB, or ΔnrpΔcydAB + nrp treated with 25 μM TTM. Error bars are SEM. Statistical significance between WT and ΔnrpΔcydAB treated with 25 μM TTM determined via two-way ANOVA with Tukey correction for multiple comparisons. ****p<0.0001. (E) Cellular ATP levels determined by BacTiter-Glo in the indicated strains treated with DMSO, 20 or 40 μM TTM, or 100 nM Q203. [ATP] determined by standard curve determined in growth media containing the same quantities of DMSO, TTM, or Q203. Error bars are SEM. Statistical significance determined via two-way ANOVA with Tukey correction for multiple comparisons. *p<0.05, ***p<0.001, ****p<0.0001. (F) Relative abundance of a CtaD-ALFA protein in M. tuberculosis of the indicated genotype treated with bathocuproinedisulfonic acid (BCS) or TTM. See Figure 3—figure supplement 1 for primary immunoblot data. Error bars are SEM. Statistical significance determined via two-way ANOVA with Tukey correction for multiple comparisons. ns = not significant.
-
Figure 3—source data 1
Raw data for Figure 3B–F.
- https://cdn.elifesciences.org/articles/105794/elife-105794-fig3-data1-v1.xlsx
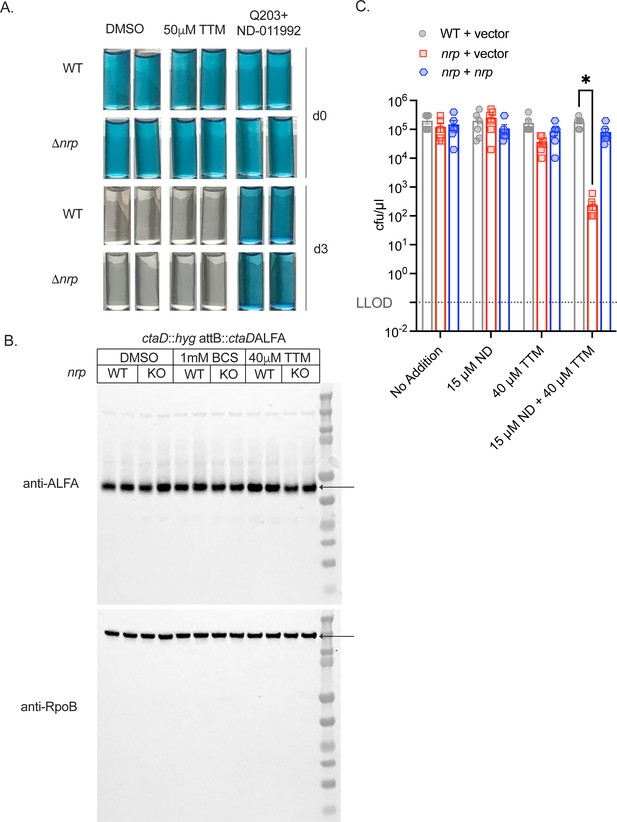
Chalkophores protect the heme-copper respiratory oxidase in copper-limiting conditions.
(A) Methylene blue decolorization assay in WT M. tuberculosis or M. tuberculosisΔnrp at assay start (d0) or after 3 d of incubation in a sealed tube (d3) treated with DMSO, 50 µM tetrathiomolybdate (TTM), or the combination of Q203 (400 nM) and ND-011992 (50 µM). (B) Copper chelation does not destabilize the CtaD protein M. tuberculosis lacking the CtaD subunit of the bcc:aa3 oxidase and complemented with a fully functional CtaD with a C-terminal ALFA epitope tag (see Figure 2E) either in the wild-type background (WT) or M. tuberculosis Δnrp (KO), which lacks diisonitrile chalkophore biosynthesis, and treated with either bathocuproinedisulfonic acid (BCS) or TTM. Full immunoblots for the ALFA tag or RpoB as a loading control are shown. Images were quantified using ImageJ software. Copper chelation synergizes with CytBD inhibition in the absence of chalkophore biosynthesis. Serial dilutions of M. tuberculosis WT,!1nrp,!1nrp +nrp strains were cultured on agar media containing TTM, ND-011992 (ND), or both. *p=0.0148 by two-way ANOVA.
-
Figure 3—figure supplement 1—source data 1
PDF file containing original western blots for Figure 3—figure supplement 1B, with bands labeled.
- https://cdn.elifesciences.org/articles/105794/elife-105794-fig3-figsupp1-data1-v1.zip
-
Figure 3—figure supplement 1—source data 2
Original uncropped western blot files for Figure 3—figure supplement 1B.
- https://cdn.elifesciences.org/articles/105794/elife-105794-fig3-figsupp1-data2-v1.zip
-
Figure 3—figure supplement 1—source data 3
Bacterial counts for Figure 3—figure supplement 1C.
- https://cdn.elifesciences.org/articles/105794/elife-105794-fig3-figsupp1-data3-v1.xlsx
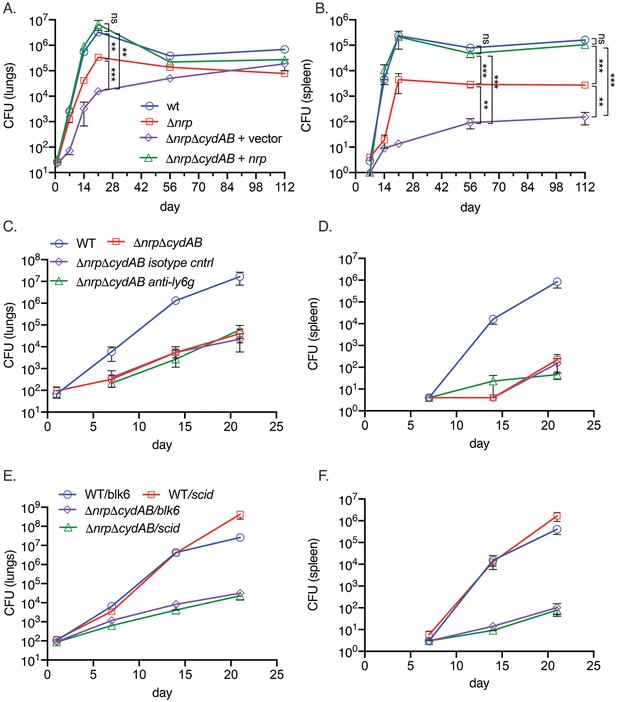
Respiratory chain flexibility is critical for M. tuberculosis virulence.
(A, B) Bacterial titers in the lung (A) or spleen (B) in mice infected with M. tuberculosis wild-type (WT), Δnrp, ΔnrpΔcydAB, or ΔnrpΔcydAB + nrp. Error bars are SEM. Statistical significance determined via two-way ANOVA with Tukey correction for multiple comparisons. Not significant (ns), **p<0.01, and ***p<0.001. (C, D) Copper deprivation by the host is independent of neutrophils. Bacterial titers in the lung (C) or spleen (D) in mice infected with M. tuberculosis WT or ΔnrpΔcydAB, or ΔnrpΔcydAB + nrp treated with isotype control antibodies or anti-Ly6G antibodies to deplete neutrophils. Flow cytometric quantitation of neutrophil depletion is provided in Figure S5. Error bars are SEM. (E, F) Copper deprivation by the host is independent of adaptive immunity. Bacterial titers in the lung (E) or spleen (F) in C57BL/6 J or C57BL/6 SCID mice infected with M. tuberculosis WT or ΔnrpΔcydAB. Error bars are SEM.
-
Figure 4—source data 1
Bacterial counts from mouse organs plotted in Figure 4A–F.
- https://cdn.elifesciences.org/articles/105794/elife-105794-fig4-data1-v1.xlsx
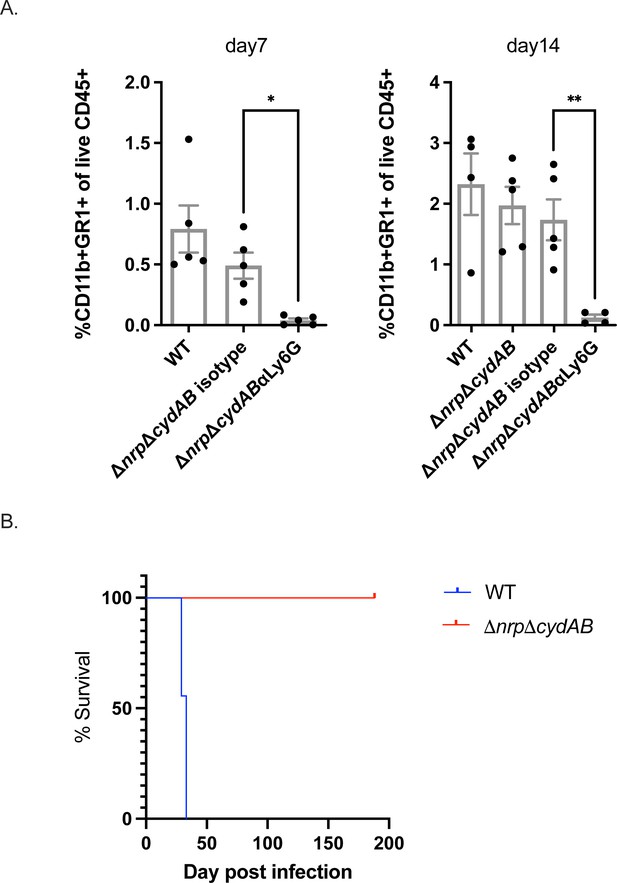
Attenuation of chalkophore-deficient M. tuberculosis is independent of neutrophils and adaptive immunity.
(A) Flow cytometric quantitation of neutrophils in the lung at day 7 and 14 post-infection with the indicated M. tuberculosis strains (wild-type, WT or ΔnrpΔcydAB) and treated with isotype control antibody or anti-Ly6G. Neutrophils were defined as CD11B+GR1+ and their percentage among CD45+ cells is graphed. Statistical significance determined by unpaired Welch’s t-test. *p<0.05, **p<0.01. Error bars are SEM (B) Survival curve of C57BL/6 SCID mice infected with WT or ΔnrpΔcydAB M. tuberculosis.
-
Figure 4—figure supplement 1—source data 1
Raw data for Figure 4—figure supplement 1A and B.
- https://cdn.elifesciences.org/articles/105794/elife-105794-fig4-figsupp1-data1-v1.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (M. tuberculosis) | nrp | erdman_0118 | ||
| Gene (M. tuberculosis) | cydA | erdman_1783 | ||
| Gene (M. tuberculosis) | cydB | erdman_1782 | ||
| Gene (M. tuberculosis) | cydD | erdman_1781 | ||
| Gene (M. tuberculosis) | cydC | erdman_1780 | ||
| Gene (M. tuberculosis) | ctaD | erdman_3330 | ||
| Gene (M. tuberculosis) | fadD10 | erdman_0116 | ||
| Strain (Escherichia coli) | DH5α | Lab Stock | ATCC SCC2197 | Plasmid Maintenance Strain |
| Strain (Escherichia coli) | EL350/pHAE87 | Lab Stock | Phage packaging strain | |
| Strain (Mus musculus) Female | B6.Cg-Prkdc(scid)/SzJ | Jackson Laboratory | Stock No 001913 | |
| Strain (Mus musculus) Female | c56bl 6 | Jackson Laboratory | Stock No 000664 | |
| Strain (M. tuberculosis) | M.tb Erdman (WT, EG2) | Lab Stock | ATCC 35801 | Animal Passaged |
| genetic reagent (M. tuberculosis) nrp KO | nrp::hygR | buglino et al. 2022 | Δnrp | Chromosomal Deletion of nt 21–7515 of Edman_0118 by double crossover recombination |
| Genetic reagent (M. tuberculosis) cydAB KO | cydAB::hygR | This Study | ΔcydAB | Chromosomal deletion of nt1 of erdman_1783 to post erdman_1782 stop codon by double crossover recombinaiton |
| Genetic reagent (M. tuberculosis) fadD10 KO | fadD10::hygR | This Study | ΔfadD10 | Chromosomal deletin of nt 85–1552 of erdman_0116 by double crossover recombination |
| Genetic reagent (M. tuberculosis) ctaD KO | ctaD::hygR | This Study | ΔctaD | Chromosomal deletion of nt 115–1723 of erdman_3330 by double crossover recombination |
| Genetic reagent (M. tuberculosis) nrp cydAB KO | nrp::loxP cydAB::hygR | This Study | ΔnrpΔcydAB | |
| Genetic reagent (M. tuberculosis) fadD10 cydAB KO | fadD10::loxP cydAB::hygR | This Study | ΔfadD10ΔcydAB | |
| Genetic reagent (M. tuberculosis) nrp ctaD KO | nrp::loxP ctaD::hygR | This Study | ΔnrpΔctaD | |
| Transfected construct (M. tuberculosis) vector | pMV306 | Lab Stock | +vector | L5 attP intergrating mycobacterium plasmid |
| Transfected construct (M. tuberculosis) nrp | pJAB823 | buglino et al. 2022 | +nrp | L5 attP integrated: AA 1–2512 erdman_0118 HSP60 promoter |
| Transfected construct (M. tuberculosis) cydABDC | pJAB900 | This Study | +cydABDC | L5 attP integrated: 330 bp 5' of erdman_1783 start to stop codon of erdman_1780 |
| Transfected construct (M. tuberculosis) ctaD | pJAB863 | This Study | +ctaD ALFA | L5 attP integrated: 310 bp 5' of erdman_3330 start to stop condon ALFA tag inserted between nt 5881 and 5882 |
| Antibody (anti-ALFA) | sdAb anti-ALFA | NanoTag Biotechnologies | N1505-HRP | 1:4000 dilution |
| Antibody (anti-RpoB) | Anti-E. coli RNA pol B | Biolegend | 663903 | 1:10000 dilution |
| Antibody (anti-mouse HRP) | IgG (H+L) Goat anti-Mouse, HRP | Fisher Scientific | 626520 | 1:10000 dilution |
| Antibody (anti-ly6G) | InVivoMAb anti-mouse Ly6G | Bio-X-Cell | BE0075-1-25MG | |
| Antibody (isotype control) | InVivoMAb rat IgG2a isotype control, anti-trinitrophenol | Bio-X-Cell | BE0089-25MG | |
| Recombinant DNA reagent | ||||
| Sequence-based reagent (qPCR primer) sigA | oSigA-1 | Intergrated DNA Technologies | 5'-cgtcttcatcccagacgaaat-3' | |
| Sequence-based reagent (qPCR primer) sigA | oSigA-2 | Intergrated DNA Technologies | 5'-cgacgaagaccacgaagac-3' | |
| Sequence-based reagent (qPCR primer) cydA | cydA FWD set3 | Intergrated DNA Technologies | 5'-gtcatcgaagtgccctatgt-3' | |
| Sequence-based reagent (qPCR primer) cydA | cydA REV set3 | Intergrated DNA Technologies | 5'-ctggtattcctgctgcagat-3' | |
| Peptide, recombinant protein | ||||
| Commercial assay or kit | In-Fusion Snap Assembly Master Mix | Takara Bio USA | 638949 | |
| Commercial assay or kit | NEBNext rRNA Depletion Kit | NEB | E785OS | |
| Commercial assay or kit | TruSeq Stranded Total RNA kit | Illumina | 20020599 | |
| Commercial assay or kit | Novaseq 6000 S4 Reagent Kit | Illumina | 20028313 | |
| Commercial assay or kit | TURBO DNA-free kit | Fisher Scientific | AM1907 | |
| Commercial assay or kit | Phusion High Fidelity Polymerase | Fisher Scientific | F530L | |
| Commercial assay or kit | GeneJET Plasmid Miniprep Kit | Fisher Scientific | FERK0503 | |
| Commercial assay or kit | Zymo Research Corporation Direct-zol RNA MiniPrep | Fisher Scientific | 50-444-622 | |
| Commercial assay or kit | Taq Universal SYBR Green Supermix | BioRad | 1725122 | |
| Commercial assay or kit | iScript cDNA Synthesis Kit | BioRad | 1708891 | |
| Commercial assay or kit | GeneJet Gel Extraction Kit | Fisher Scientific | FERK0692 | |
| Chemical compound, drug | methylene blue | Fisher Scientific | S25429 | |
| Chemical compound, drug | dimethyl sulfoxide (DMSO) | Sigma | D2650 | |
| Chemical compound, drug | Ammonium tetrathiomolybdate (TTM) | Sigma | 323446 | |
| Chemical compound, drug | Bathocuproinedisulfonic acid disodium salt (BCS) | Sigma | B1125 | |
| Chemical compound, drug | Telacebec (Q203) | AbMole BioScience | M5297 | |
| Chemical compound, drug | ND-011992 | This Study | synthesized by Tan lab, see methods | |
| Chemical compound, drug | diisonitrile | This Study See supplemental methods | ||
| Chemical compound, drug | TRIzol Reagent | Fisher Scientific | 15-596-026 | |
| Software, algorithm | fastqc | http://www.bioinformatics.babraham.ac.uk/projects/fastqc | ||
| Software, algorithm | bwa mem | Li and Durbin, 2009 | ||
| Software, algorithm | samtools | Li et al., 2009 | ||
| Software, algorithm | Bioconductor Rsubread package | Liao et al., 2014 | ||
| Software, algorithm | DESeq2 R package | Love et al., 2014 | ||
| Software, algorithm | R | The R Project for Statistical Computing | https://www.R-project.org | |
| Other | NUPAGE 4–12% BT Gel | Fisher Scientific | NPO312BOX/NPO336BOX | |
| Other | Protran Nitrocellulose Hybridization Transfer Membrane | Perkin Elmer | NBA08C001EA | |
| Other | Fibox 4 trace instrument | PreSens | https://www.presens.de/products/detail/fibox-4-trace | |
| Other | Oxygen detection sensor spot PSt6 | PreSens | https://www.presens.de/products/detail/oxygen-sensor-spot-sp-pst6-nau |
Additional files
-
Supplementary file 1
Strains used in this study.
- https://cdn.elifesciences.org/articles/105794/elife-105794-supp1-v1.xlsx
-
Supplementary file 2
Plasmids used in this study.
- https://cdn.elifesciences.org/articles/105794/elife-105794-supp2-v1.xlsx
-
Supplementary file 3
RNA sequencing dataset.
- https://cdn.elifesciences.org/articles/105794/elife-105794-supp3-v1.xlsx
-
Supplementary file 4
Chemical synthesis methods, 1H NMR, and 13C NMR spectra.
- https://cdn.elifesciences.org/articles/105794/elife-105794-supp4-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/105794/elife-105794-mdarchecklist1-v1.docx





