Functional analysis across model systems implicates ribosomal proteins in growth and proliferation defects associated with hypoplastic left heart syndrome
Figures

Whole-genome siRNA screen identified ribosomal proteins as agonists of cardiomyocyte (CM) proliferation.
(A) High-throughput iPSC-derived CM proliferation screen overview. (B) Screen result showing normalized % EdU+ CMs (X-axis) and relative total number of CMs (Y-axis) upon knockdown of genome-wide siRNAs (18,055 siRNAs). siRNAs for RPL and RPS genes highlighted in red. (C) Representative immunofluorescence images of proliferation (EdU incorporation, green, CM marker ACTN2, red) of induced hPSC-CMs upon TP53 and RPS14 knockdown. Insets: nuclei (DAPI). (D) Gene ontology enrichment analysis for whole-genome sequencing (WGS) hits (BP, biological process; FDR-corrected analysis using gprofiler2). (E) Overview of hits corresponding to top 4 non-redundant BP categories.
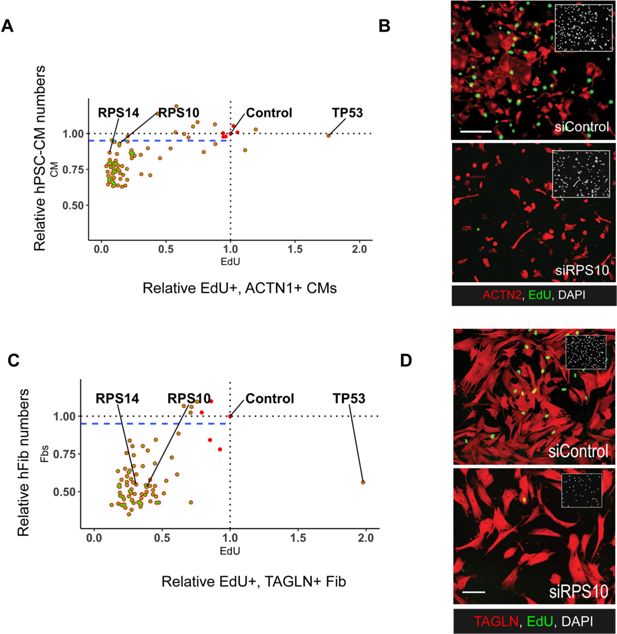
Functional validation of RP–mediated control of proliferation in hiPSC-CMs and human dermal fibroblasts.
(A, B) Confirmatory siRNA screening for all RP genes confirms critical role for RPs in cardiomyocyte (CM) proliferation. (C, D) KD of RPs reduces proliferation in human fibroblasts.

Ribosomal gene variants identified in hypoplastic left heart syndrome (HLHS).
(A) Gene prioritization scheme of 25 poor-outcome proband–parent trios. (B) Testing 292 HLHS candidate genes from all poor-outcome families in cardiomyocytes (CMs) identified RPs as major regulators of hPSC-CM proliferation (normalized fraction of ACTN2+/EdU+ cells). (C) Drosophila cardiac phenotypes induced by loss of RP genes affected in HLHS patients with poor outcome. The heart is visualized by RFP expression specifically in CMs (R94C02::tdTom). Knockdown is achieved by sustained Gal4/UAS activity using Hand4.2-Gal4. (D) Wild-type zebrafish larva, rps17 and rps28 CRISPR mutants, and rpl39 morphants at 72 hpf. rpl39 morphants, injected with 1 ng MO in lateral view, show mild edema. (E) Systolic surface area (SA) upon rps17 and rps28 CRISPR, and diastolic SA after rpl39 MO in the atrium and ventricle of zebrafish hearts.

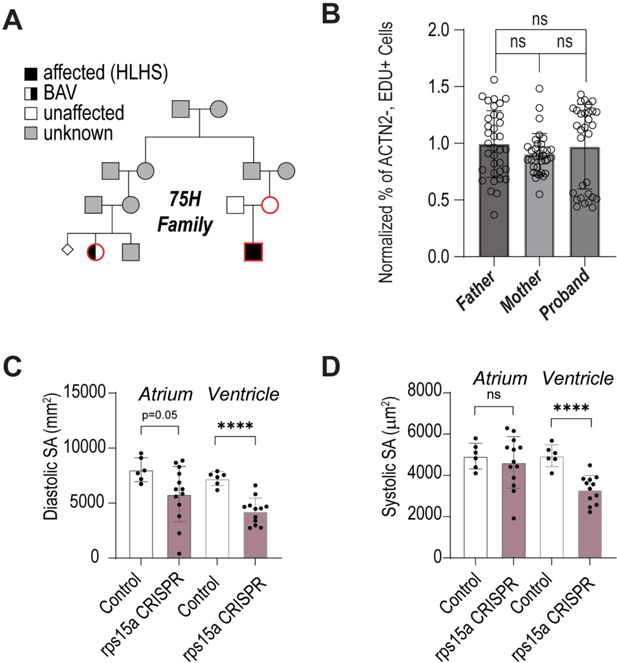
Characterization of RPS15A from the 75H hypoplastic left heart syndrome (HLHS) index family.
(A–C) Prioritized candidate genes from the 75H family and relative hPSC-cardiomyocyte (CM) proliferation capacity upon KD. (D) Representative immunofluorescence images of proliferation (EdU incorporation, green; CM marker ACTN2, red) of induced hPSC-CMs upon siRPS15A knockdown. (E) Heart-specific KD of RpS15Aa in Drosophila adult hearts causes loss of heart tissue and is fully penetrant. (F) Representative immunofluorescence images of control and RpS15Aa-RNAi show partial heart loss (myosin heavy chain, Mhc, red; heart tissue-reporter, green). (G) CRISPR-mediated loss of rps15a in F0 larval zebrafish hearts causes decrease in CM number. (H) Representative immunofluorescence of hearts and whole-mount images of control and rps15a-CRISPR F0 larval hearts (CM nuclei reporter, red).
Model organism phenotypes caused by loss of RP genes.
(A) Pedigree of the extended 75H family investigated for shared variants between hypoplastic left heart syndrome (HLHS) proband (black square) and distant cousin with congenital heart disease (CHD) (half-filled circle). (B, C) Diastolic and (D) systolic surface area after rps15a CRISPR. Statistics: Mann–Whitney test, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
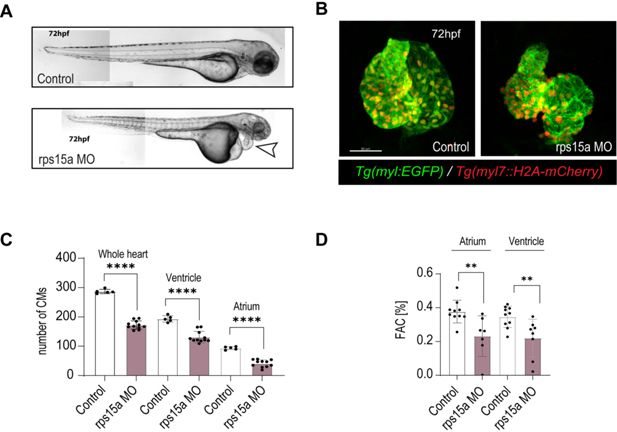
rps15a zebrafish larval morphants show heart dysfunction and reduction in cardiomyocyte (CM) number.
(A) Lateral view (head to the right) of a wild-type control zebrafish larva (top) and a morphant (bottom) following injection of 2 ng/µl rps15a morpholino (MO). The morphant exhibits near-normal body/tail but notable pericardial edema (arrowhead). (B) Control (left) and morphant hearts (right) stained with Tg(myl7:EGFP) and Tg(myl7:H2A-mCherry). Note the smaller heart with aberrant looping in morphant heart. (C) Total CM counts (mCherry+ cells) in control and rps15a morphants shown in (B). (D) Fractional area change (FAC) in control and rps15a morphants. Student’s t-test, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
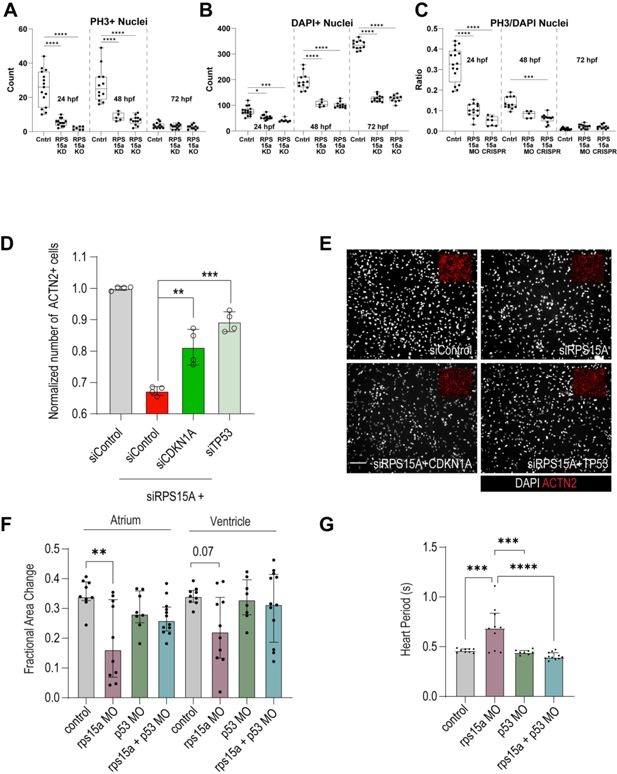
Proliferation and heart function depend on RPS15A and are regulated via TP53 in cardiomyocytes (CMs) and zebrafish.
Cardiac proliferation (by PH3, A), total CM number (DAPI, B) and ratio of proliferating CMs (C) assessed for rps15a morphants (KD) and CRISPR (KO) for three timepoints, 24/48/72 hpf (D, E) The number of siRPS15A-treated CMs is strongly reduced, which can be attenuated by co-KD of CDKN1A or TP53. (F, G) Loss of rps15a causes strong reduction in zebrafish heart contractility (measured by fractional area change, C) and heart period (D), which can be rescued by co-KD of p53 in both zebrafish heart atrium and ventricle.

Loss of ribosomal gene function in cardiomyocytes (CMs) invokes TP53-stress response.
(A) RNA-sequencing of hPSC-CMs following siRNA treatment for RPL39 and RPS15A shows both convergent and divergent transcriptomic response. (B) Gene ontology (GO) term analysis of differentially expressed genes following RP KD shows TP53-mediated response, including upregulation of CDKN1A. (C, D) CDKN1A is highly upregulated in CMs following siRPS15A treatment. (E, F) Reduced CM proliferation upon RPS15A KD is mediated by CDKN1A/TP53 and can be rescued upon their co-KD. (G, H) Larval zebrafish CM number is reduced by morpholino treatment for rps15a and can be attenuated by P53 morpholino co-injection. Control and morphant (MO) hearts of 72 hpf zebrafish larva stained with Tg(myl7:EGFP) and Tg(myl7:H2A-mCherry). Note that the smaller heart with aberrant looping by rps15a MO is partially reversed by p53 co-KD. Student’s t-test, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.

Rescue of RpS15Aa KD-mediated heart tube loss in Drosophila by YAP/yorkie overexpression depending on its co-factor TEAD/scalloped.
(A) Representative images of RFP-expressing fly hearts. RpS15Aa KD-mediated heart tube loss can be partially rescued by overexpression of yorkie (RpS15Aa RNAi + yorkie OE). The rescue by yki OE depends on its co-factor sd. Flies were raised at 25°C. (B) Quantification of events presented as a percentage of flies exhibiting whole heart tube versus partial heart loss (defined as 25–75% heart tube length compared to wildtype) or no heart tube. Statistics: Fisher’s exact test, *p < 0.05. (C) Proposed signaling cascade underlying cardiac growth, proliferation, and differentiation impairment following ribosomal stress (adapted from Baker et al., 2019).

RPS15A genetically interacts with cardiac transcription factors.
(A) The majority of cardiac transcription factors do not impact cardiomyocyte (CM) proliferation, except for, e.g., HES4 and HOPX. (B) TBX5 genetically interacts with RPS15A and RPL39. siTBX5 does not impact CM proliferation at 0.5, 1, 1.5, or 2 nM si-concentration, and neither do siRPS15A and siRPL39 alone at 0.5 nM. CM proliferation is reduced in siRP backgrounds with increased titration of siTBX5. Two-way ANOVA for siTBX5 dosage, RP-knockdown, and their interaction. (C) Representative fly heart segment (A4) from control flies, heterozygous mutants (tin346/+, pnrVX6/+, DocDf/-, Df(RpS15Aa)+/-) and transheterozygous mutants. tin+/- = tin346/+, pnr+/- = Df(pnr)/+, Doc+/- = Df(DocA)/+. Note the deformation and myofibrillar disorganization in the transheterozygous mutants. (D) Quantification of adult Drosophila heart defects and genetic interaction. Statistics: Fisher’s exact test on absolute numbers testing normal versus severely deformed hearts. *p < 0.05, **p < 0.005, ***p < 0.001.
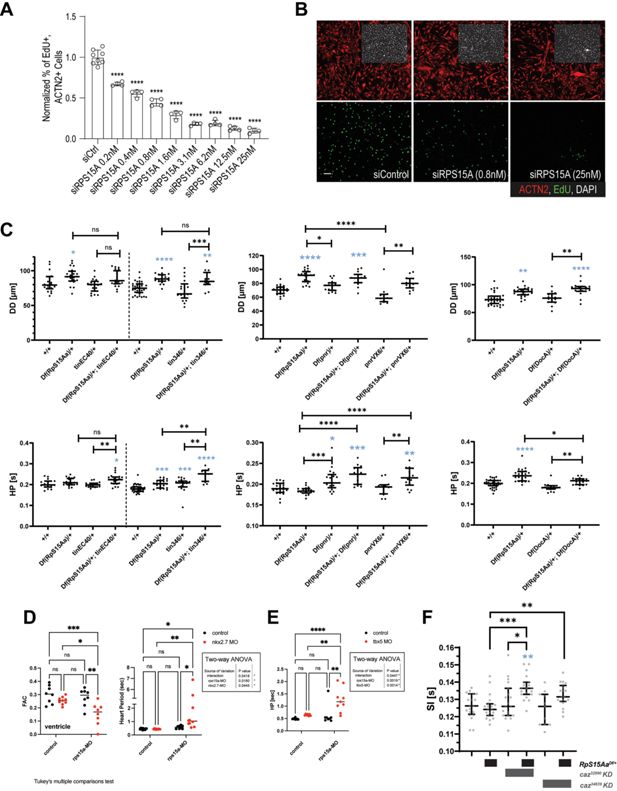
Genetic interaction between RpS15Aa/RPS15A and cabeza/EWSR1.
(A, B) Dose-dependent inhibition of hPSC-cardiomyocytes (CMs) proliferation by RPS15A KD. (C) Diastolic diameter (DD), and heart period (HP) of 3-week-old female control and heterozygous RpS15Aa deficient flies with or without additional heterozygous mutation in tinman, pannier, or Dorsocross. Statistics: Kruskal–Wallis, *p < 0.05, **p < 0.005, ***p < 0.001, ****p < 0.0001. Note the prolonged HP in transheterozygous mutants for Df(RpS15Aa) and tinman. (D, E) FAC and HP of the atria and ventricles at 72 hpf in zebrafish embryos injected with 0.5 ng rps15a and/or 1 ng nkx2.7 MOs (E) or 0.5 ng rps15a and/or 0.5 ng tbx5 MOs (F). Note that FAC and heart periods show synergistic genetic interaction between rps15a and nkx2.7 and tbx5a. Statistics: Two-way ANOVA, *p < 0.05, **p < 0.01, ****p < 0.0001. (F) Systolic intervals (SI) of 1-week-old adult flies upon heart-specific knockdown of cabeza (caz32990 or caz34839 RNAi) using Hand4.2-Gal4 driver with or without additional RpS15Aa heterozygosity (RpS15AaDf/+) and age-matched controls. Automated analysis of SI depicted as median with interquartile range obtained from 5 s high-frame-rate videos. Statistics: Kruskal–Wallis test, *p < 0.05, **p < 0.01, ***p < 0.001.

RP-dependent cardiac-specific regulation of cell proliferation.
(A) Schematic illustrating approach to identify novel RP-dependent and cardiac-specific hypoplastic left heart syndrome (HLHS)-associated gene network controlling cardiomyocyte (CM) proliferation. (B) Heatmap showing differential expression (hiPSC-CMs vs hiPSCs) of genes regulating CM proliferation. (C) Histogram showing effect of cardiac-specific and HLHS-associated genes on CM proliferation. (D) Visualization of RP-dependent and cardiac-specific HLHS-associated gene network (GeneMania). HLHS families. (E) Table of HLHS families harboring rare and damaging variant in RP-dependent and cardiac-specific genes. (F) Histogram showing lack of effect of siEWSR1 on OCT4+ hiPSCs. (G) Representative images of OCT4+ cells in siControl and siEWSR1. (H) Histogram showing that siEWSR1 increases the % of CDKN1A+ CMs as compared to siControl. (I) Histogram showing that siEWSR1 concomitantly decreases the % of EDU+ CMs as compared to siControl. (J) Representative images showing immunostaining for CDKN1A (white), EDU (green), and ACTN2 (red) in siEWSR1 and siControl conditions. (K) Pathway reconstruction of cardiac and RP-dependent regulation of CM proliferation by HLHS-associated genes. Chi-square test, *p < 0.05, **p < 0.005, ***p < 0.001.

Model for RP-dependent involvement in hypoplastic left heart syndrome (HLHS)-associated phenotypes.
Schematic showing that combined prioritization and unbiased screening led to the identification of a novel HLHS-associated gene network regulating cardiomyocyte (CM) proliferation as potential disease-causing mechanism.
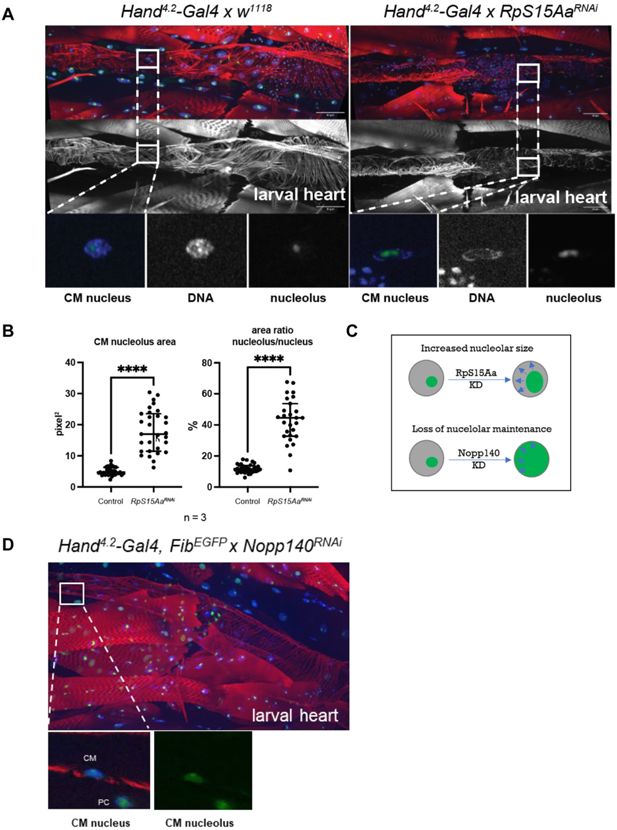
RpS15Aa KD induces nucleolar stress in third instar larval hearts.
(A) Nucleolar stress induction in third instar larval hearts upon RpS15Aa KD as indicated by enlarged nucleoli. Hearts from control and RpS15Aa KD. Third instar larva stained for F-actin (red), DAPI (blue), and nucleolus marker Fibrillarin (green) and cardiomyocyte (CM) nuclei in magnifications. Note, heart constriction upon RpS15Aa KD in the larval heart. (B) Quantification of nucleolus area and area ratio (nucleolus/nucleus) indicates significantly enlarged nucleoli upon RpS15Aa KD. Statistics: Mann–Whitney test, ****p < 0001. (C) Schematic representation of different nucleolar phenotypes upon RNAi-mediated loss of function. (D) As a positive control, KD of another nucleolar key player Nopp140 specifically in the heart using Hand4.2-Gal4 combined with FibrillarinEGFP (kind gift of the Wieschaus lab) leads to loss of nucleolar maintenance. Third instar larva stained for F-actin (red) and DAPI (blue). Fibrillarin in green. CM nuclei in magnifications. PC = pericardial cell.
Tables
Variants in ribosomal genes in hypoplastic left heart syndrome (HLHS) probands.
| Gene | Proband (age, sex) | Mode of inheritance | Variant | Type | MAF% | CADD score | TFBS affected | hPSC-CM proliferation | Fly gene and defects | Zebrafish gene and defects | Patient outcome |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RPL26L1 | 145H (3y, m) | Compound heterozygous | –1248A>G; V97M | Regulatory; missense | 0.029; 0.032 | -; 21.3 | Pdx1; NFE2L1::MAFG; FOXC1 | Reduced | RpL26: lethal, no heart | rpl26: n.t. | Restrictive ASD, PLE |
| RPL36A | X-linked recessive | –1321C>T | Regulatory | 0.055 | - | PAX2 | No effect | RpL36A: lethal, no heart | rpl36a: n.t. | ||
| RPS15 | Compound heterozygous | –1558C>T; T101S | Regulatory; missense | 0; 0.102 | -; 23.5 | FOXC1 | Reduced | RpS15: lethal | rps15: n.t. | ||
| RPL39 | 151H (20y, m) | X-linked recessive | –1359T>C | Regulatory | 0.653 | - | HOXA5 | Reduced | RpL39: lethal | rpl39: morphants mild edema, reduced ventricular size | Reduced RV function |
| RPL3L | 96H (22m, m) | Compound heterozygous | R200Q; R242W | Missense; missense | 0.966; 0.432 | 21.2; 14.94 | - | Elevated | RpL3: no heart | rpl3: n.t. | Reduced RV function |
| RPL13A | 201H | Compound heterozygous | –92–645C>T; –29–191C>T | Regulatory; regulatory | 0.72; 0.046 | - | FOXD1; GATA2; ETS1; ELF5; FOXC1 | No effect | RpL13A: no phenotype | rpl13a: n.t. | Failing Fontan circulation, transplant at 14y |
| RPS17 | 325H | Homozygous recessive | S136N | Missense | 0 | <10 | - | Reduced | RpS17: lethal | rps17: CRISPR mutants show systolic atrial dysfunction, shortened heart period | Reduced RVEF and increased RVEDP at 9m |
| RPL10 | 76H | X-linked recessive | 24–218G>A | Regulatory | 0.583 | - | ELK1; ETS1; SPIB; POLR2A; HEY1; Hltf | Reduced | RpL10: constricted | rpl10: n.t. | Reduced RVEF at 9y |
| RPS28 | Compound heterozygous | –589G>A; –505A>T | Regulatory; regulatory | 0.061; 0.08 | - | CTCF | Reduced | RpS28b: ostia defect | rps28: CRISPR mutants show no heart phenotype |
-
MAF – minor allele frequency; TFBS – transcription factor-binding site; n.t. – not tested; ASD – atrial septal defect; PLE – protein-losing enteropathy; RV – right ventricle; RVEF – right ventricular ejection fraction; RVEDP – right ventricular end diastolic pressure.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (D. melanogaster) | Hand4.2-Gal4 | Bodmer lab | PMID:16467358 | |
| Genetic reagent (D. melanogaster) | R94C02::tdTomato | N. Jan lab | FBtp0137272 | |
| Genetic reagent (D. melanogaster) | UAS-RpS15AaRNAi | Vienna Drosophila Resource Center (VDRC) | FBgn0010198 | v19198 |
| Genetic reagent (D. melanogaster) | Df(RpS15Aa) | Bloomington Drosophila Stock Center (BDSC) | FBab0047266 | 39614 |
| Genetic reagent (D. melanogaster) | UAS-RpL26RNAi | Vienna Drosophila Resource Center (VDRC) | FBgn0036825 | v40402 v100280 |
| Genetic reagent (D. melanogaster) | UAS-RpL36ARNAi | Vienna Drosophila Resource Center (VDRC) | FBgn0031980 | v108391 |
| Genetic reagent (D. melanogaster) | UAS-RpS15RNAi | Vienna Drosophila Resource Center (VDRC) | FBgn0034138 | v35415 v104439 |
| Genetic reagent (D. melanogaster) | UAS-RpL39RNAi | Vienna Drosophila Resource Center (VDRC) | FBgn0023170 | v23578 v108821 |
| Genetic reagent (D. melanogaster) | UAS-RpL3RNAi | Vienna Drosophila Resource Center (VDRC) | FBgn0020910 | v109820 |
| Genetic reagent (D. melanogaster) | UAS-yorkie | Pan | ||
| Genetic reagent (D. melanogaster) | UAS-MycRNAi | Bloomington Drosophila Stock Center (BDSC) | FBgn0262656 | 25784 |
| Genetic reagent (D. melanogaster) | UAS-sdRNAi | Vienna Drosophila Resource Center (VDRC) | FBgn0003345 | v101497 |
| Genetic reagent (D. melanogaster) | Df(3L)DocA | Reim | Fbab0037663 | |
| Genetic reagent (D. melanogaster) | tinEC40 | Bloomington Drosophila Stock Center (BDSC) | Fbal0032861 | 78560 |
| Genetic reagent (D. melanogaster) | tin346 | Bloomington Drosophila Stock Center (BDSC) | Fbal0035787 | 92964 |
| Genetic reagent (D. melanogaster) | pnrVX6 | Bloomington Drosophila Stock Center (BDSC) | Fbal0032468 | 6334 |
| Genetic reagent (D. melanogaster) | Df(pnr) | Bloomington Drosophila Stock Center (BDSC) | Fbab0038315 | 7982 |
| Strain, strain background (Danio rerio) | Oregon AB wild-type | Ocorr lab, SBP | A commonly used wild-type strain | |
| Strain, strain background (Danio rerio) | Tg(myl7:EGFP)twu277 | Tsai Lab, National Taiwan University | PMID:12950077 | A transgenic line of zebrafish labeled with heart-specific EGFP fluorescence |
| Strain, strain background (Danio rerio) | Tg(myl7:H2A-mCherry)sd12 | Yelon Lab, University of California, San Diego | PMID:24075907 | A transgenic line of zebrafish specifically expressing mCherry in cardiomyocyte nuclei |
| Antibody | Mouse monoclonal anti-ACTN1 | Sigma | A7811 | 1:800 |
| Antibody | Mouse monoclonal anti-POU5F1 (OCT4) | Sigma | P0082 | 1:500 |
| Antibody | Donkey polyclonal anti-mouse Alexa Fluor 568 | Invitrogen | A10037 | 1:500 |
| Antibody | Chicken polyclonal anti-GFP | Aves Labs | GFP-1020 | 1:300 |
| Antibody | Rabbit polyclonal anti-mCherry | Rockland | 600-401P16S | 1:200 |
| Antibody | Donkey polyclonal anti-chicken AlexaFluor 488 | Jackson ImmunoResearch | 703-545-155 | 1:200 |
| Antibody | Donkey polyclonal anti-rabbit AlexaFluor 568 | Invitrogen | A10042 | 1:200 |
| Other | DAPI (iPSC) 500 mg/ml | Sigma | D9542 | Nuclear stain 1:1000 |
| Antibody | Mouse anti-Mhc (Drosophila) | DSHB | 3E8-3D3 | 1:50 |
| Antibody | Anti-mouse-Alexa Fluor 488 | Jackson Labs | 115-545-003 | 1:500 |
| Antibody | Alexa Fluor 647 phalloidin | Invitrogen | A22287 | 1:500 |
| Other | DAPI (Zebrafish) 500 mg/ml | Invitrogen | D1306 | Nuclear stain 1:200 |
| Sequence-based reagent | RPS15A siRNA | Entrez Gene ID: 6210 | Dharmacon | On-Target plus, individual sequence |
| Sequence-based reagent | TP53 siRNA | Entrez Gene ID: 7157 | Dharmacon | On-Target plus, Individual Sequence |
| Sequence-based reagent | CDKN1A siRNA | Entrez Gene ID: 1026 | Dharmacon | On-Target plus, SmartPool |
| Sequence-based reagent | TP53 | 371502118c1 | IDT Integrated DNA Technologies, Coralville, IA | Expression level |
| Sequence-based reagent | CDKN1A | 310832423c1 | IDT Integrated DNA Technologies, Coralville, IA | Expression level |
| Sequence-based reagent | CCNB1 | 356582356c1 | IDT Integrated DNA technologies, Coralville, IA | Expression level |
| Sequence-based reagent | CCNB2 | 332205979c1 | IDT Integrated DNA technologies, Coralville, IA | Expression level |
| Sequence-based reagent | CDK1 | 281427275c1 | IDT Integrated DNA technologies, Coralville, IA | Expression level |
| Sequence-based reagent | MCM2 | 33356546c1 | IDT Integrated DNA technologies, Coralville, IA | Expression level |
| Sequence-based reagent | RPS15A | 71772358c2 | IDT Integrated DNA Technologies, Coralville, IA | Expression level |
| Sequence-based reagent | GAPDH | Hs.PT.45.8326 | IDT Integrated DNA Technologies, Coralville, IA | Expression level |
| Commercial assay or kit | EdU | Click-it Plus EdU Imaging Kit | Life Technologies | |
| Software, algorithm | Prism v7 and v8 | SBP license | GraphPad Software |
Additional files
-
Supplementary file 1
Whole-genome siRNA screen results table.
- https://cdn.elifesciences.org/articles/106231/elife-106231-supp1-v1.xlsx
-
Supplementary file 2
292 filtered variants from poor-outcome HLHS probands.
MOI – mode of inheritance.
- https://cdn.elifesciences.org/articles/106231/elife-106231-supp2-v1.xlsx
-
Supplementary file 3
Subset from all 292 genes that cause CM proliferation defects and/or fly heart defects.
- https://cdn.elifesciences.org/articles/106231/elife-106231-supp3-v1.xlsx
-
Supplementary file 4
Segregating variants of 75H proband.
- https://cdn.elifesciences.org/articles/106231/elife-106231-supp4-v1.xlsx
-
Supplementary file 5
Differential gene expression lists for siRPL39 and siRPS15A in hPSCs and CMs.
- https://cdn.elifesciences.org/articles/106231/elife-106231-supp5-v1.txt
-
Supplementary file 6
Differential gene expression lists for siRPL39 and siRPS15A in hPSCs and CMs.
- https://cdn.elifesciences.org/articles/106231/elife-106231-supp6-v1.txt
-
Supplementary file 7
Differential gene expression lists for siRPL39 and siRPS15A in hPSCs and CMs.
- https://cdn.elifesciences.org/articles/106231/elife-106231-supp7-v1.txt
-
Supplementary file 8
Differential gene expression lists for siRPL39 and siRPS15A in hPSCs and CMs.
- https://cdn.elifesciences.org/articles/106231/elife-106231-supp8-v1.txt
-
Supplementary file 9
Twelve genes prioritized after RNA-seq and whole-genome sequencing (WGS) analysis.
- https://cdn.elifesciences.org/articles/106231/elife-106231-supp9-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/106231/elife-106231-mdarchecklist1-v1.docx







