The nucleolar protein NIFK promotes cancer progression via CK1α/β-catenin in metastasis and Ki-67-dependent cell proliferation
Figures
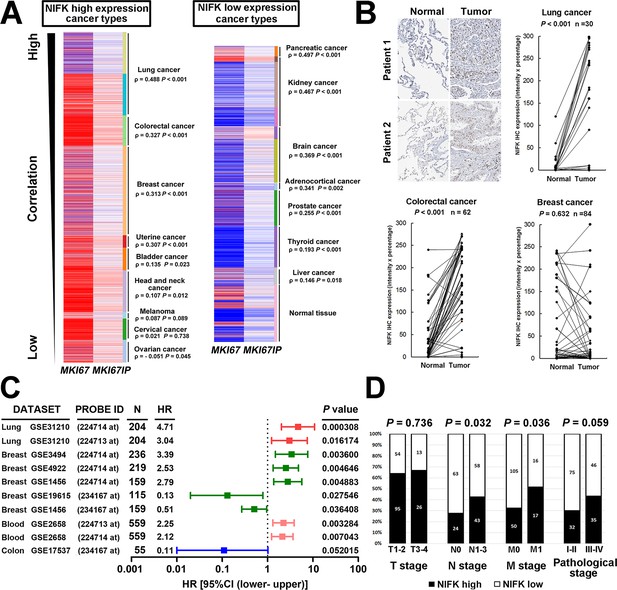
NIFK expression is most concurrently elevated with Ki67 in lung cancer and lung cancer patients displaying high NIFK level exhibit frequent lymph node and distant metastasis.
(A) In the TCGA pan-cancer cohort, significantly positive correlations between MKI67IP (NIFK) and MKI67 (Ki-67) RNA expression were observed in almost all cancer types. Among the cancer types that displayed high MKI67IP expression, lung cancer exhibited the strongest correlation between MKI67IP and MKI67 expression. Red color in heat map represents genes with high expression. Blue color in heat map represents gene with low expression. (B) Based on IHC analysis, significant overexpression of NIFK compared with paired normal tissue was observed in lung cancer (upper) and colorectal cancer (lower left) but not in breast cancer (lower right). (C) Forest plot comparison of the hazard ratio of MKI67IP (NIFK) overexpression in patients with various cancer types revealed that lung cancer displayed the strongest impact of NIFK RNA expression on survival. (D) Lung cancer patients displaying NIFK overexpression exhibited more frequent lymph node and distant metastasis and a higher pathological stage.
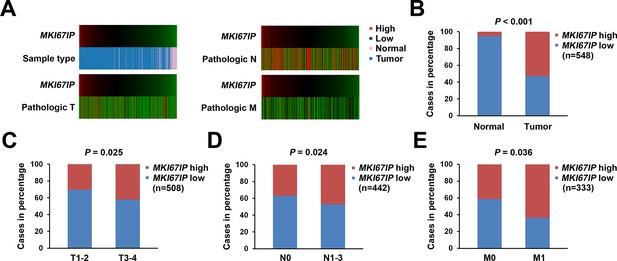
MKI67IP is overexpressed in tumors and its expression correlates with the pathological TNM stage in lung cancer.
The heat map (A) illustrates the association of MKI67IP expression with the corresponding (B) sample type, (C) T stage, (D) N stage and (E) M stage in the lung cancer cohort. The data for 548 cases were retrieved from the TCGA (lung adenocarcinoma (LUAD) gene expression by RNAseq (Illumina HiSeq)) database and were analyzed using the chi-squared test.
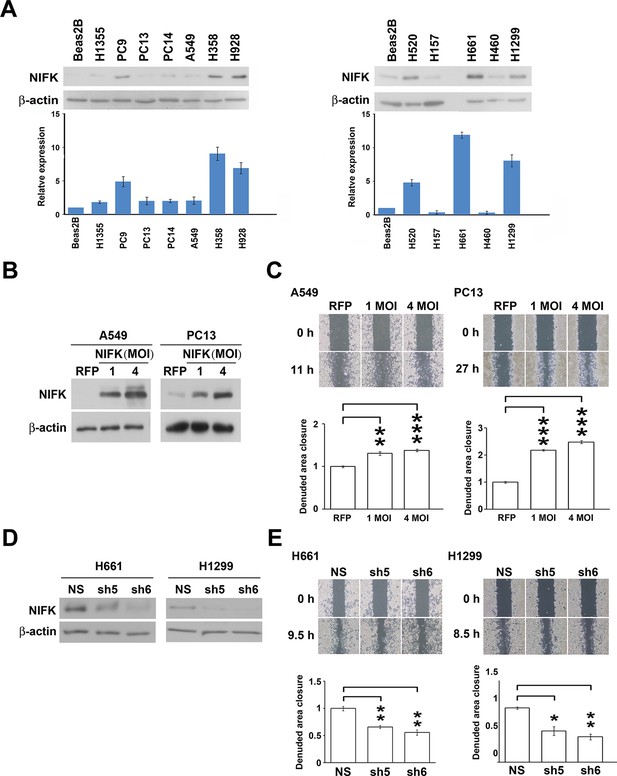
NIFK promotes the migration of lung cancer cells in vitro.
(A) The endogenous expression levels of NIFK in lung adenocarcinoma (Left), squamous and large cell lung cancer cell lines (Right). The relative expression levels were normalized to those of normal Beas2B lung cells, and the average expression levels are presented. (B) The relative NIFK levels in A549 and PC13 cells after overexpression of NIFK via lentiviral infection. (C) The migratory capacity of NIFK-overexpressing A549 and PC13 cells was assessed using a wound-healing assay. The exposed area was measured after the indicated incubation period and was normalized to that of the 0-h control. (D) The NIFK knockdown efficiencies in the lentivirus-based shRNA clones sh5 and sh6, corresponding to H661 and H1299 cells, respectively. NS, non-silenced control. (E) H661 and H1299 cell migration after NIFK knockdown was evaluated at the indicated time points.
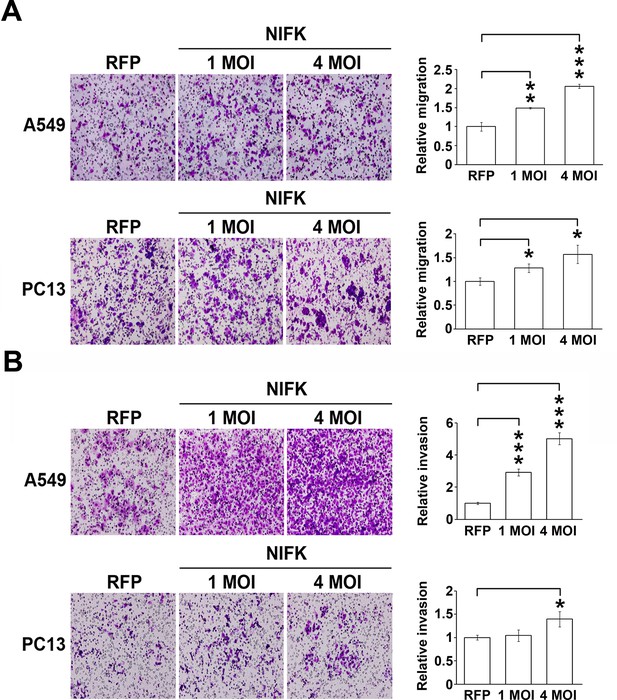
The overexpression of NIFK promotes cell migration, invasion in vitro and metastasis in vivo.
(A-B) Cell migration and invasion were evaluated via the transwell assay. A549 cells and PC13 cells were incubated for 4.5 hr and 24 hr, respectively, to evaluate cell migration. For the invasion assay, both A549 and PC13 cells were incubated for 42 hr.
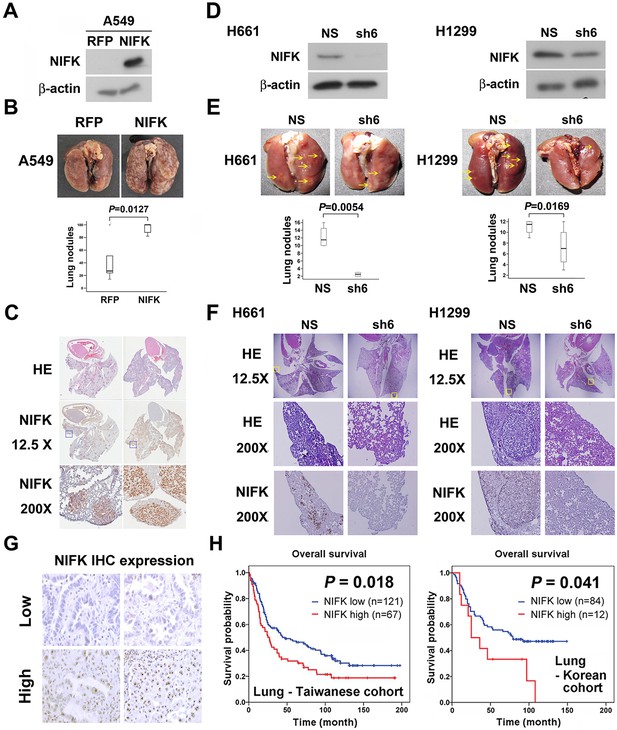
NIFK facilitates cancer cell metastasis in vivo and is associated with poor survival of lung cancer cohorts.
(A) The level of NIFK overexpression following infection of A549 cells with 4 MOI virus. (B) The indicated cells were injected into NSG mice via the tail vein. Surface lung nodules were statistically quantified. N=5 per group. (C) Representative images of surface lung nodules with HE staining and IHC staining for NIFK are presented for the RFP- or NIFK-overexpressing cell-injected groups. (D) The NIFK knockdown efficiency in H661 and H1299 cells. (E) The cells were injected via the tail vein of the mice. Top, representative images of lung metastasis of the indicated H661 and H1299 cells. Bottom, statistical quantification of lung metastatic nodules in the indicated groups. (F) Top, representative images of lung HE staining. Middle, images of HE staining in the indicated areas. Bottom, IHC staining for NIFK in the indicated areas. (G) Representative images of IHC staining for NIFK. (H) Kaplan-Meier survival analysis revealed that high NIFK IHC expression correlates with poor prognosis in lung cancer.
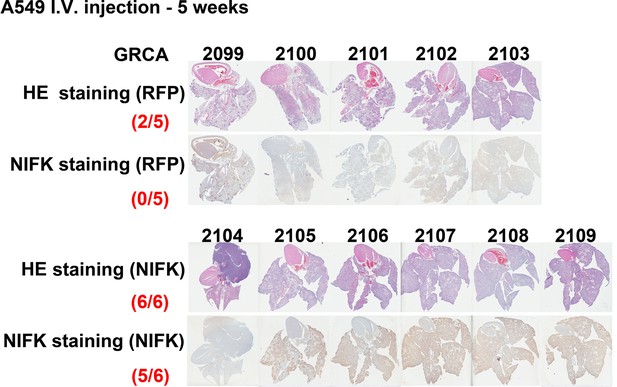
Cells overexpressing NIFK or RFP were injected via the tail vein.
After 5 weeks, the mice were sacrificed, and each mouse was assigned a GRCA number. HE and IHC staining for NIFK were performed and are presented.
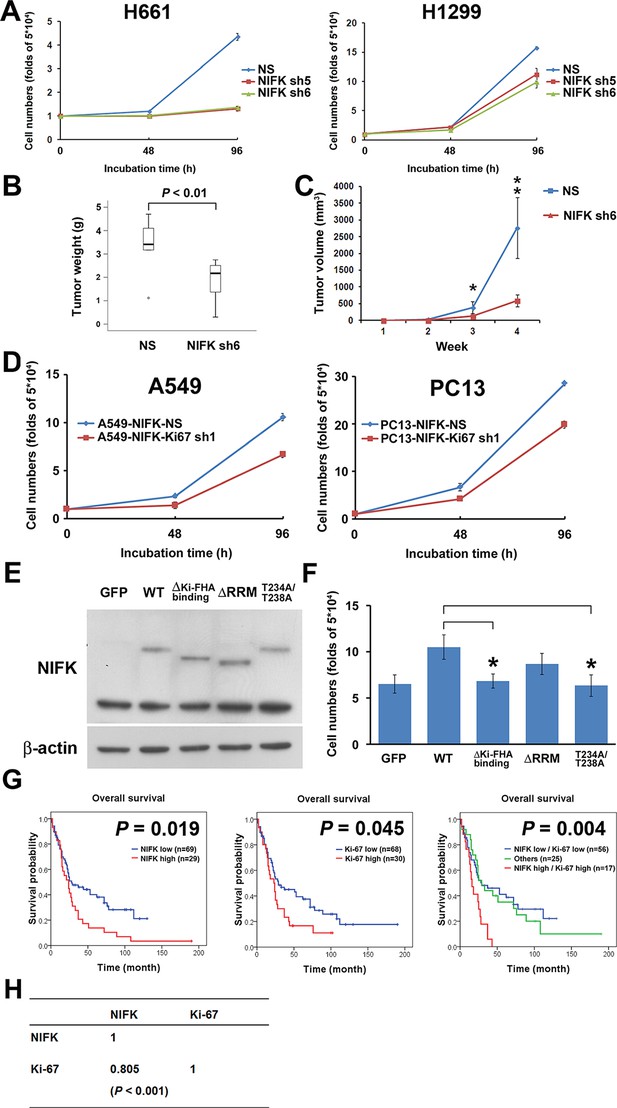
NIFK promotes cell proliferation via the poor prognosis marker Ki-67.
(A) H661 and H1299 cells were seeded at a density 5×104 cells/well. The number of cells was counted at 48 and 96 hr via the Trypan blue exclusion assay. (B) Box plot represented tumor weight of H1299 cell NS clone and NIFK-silenced clone at 4 weeks after injection. (C) Growth curves of tumor volume in indicated groups. (D) Cell numbers of NIFK overexpressing A549 and PC13 cells upon Ki-67 silence. (E) Overexpression of GFP-tagged wild type (WT), truncated and point mutated NIFK in PC13 cells. Cells were transiently transfected via liposome and selected. (F) Cell numbers of indicated groups were evaluated at 48 hr after cell seeding at density 5×104 cells/well in 6 wells plate. (G) Kaplan-Meier survival analysis revealed the correlation of NIFK and Ki-67 IHC expression with poor prognosis in lung cancer. (H) The correlation of NIFK and Ki-67 IHC expression in lung cancer.
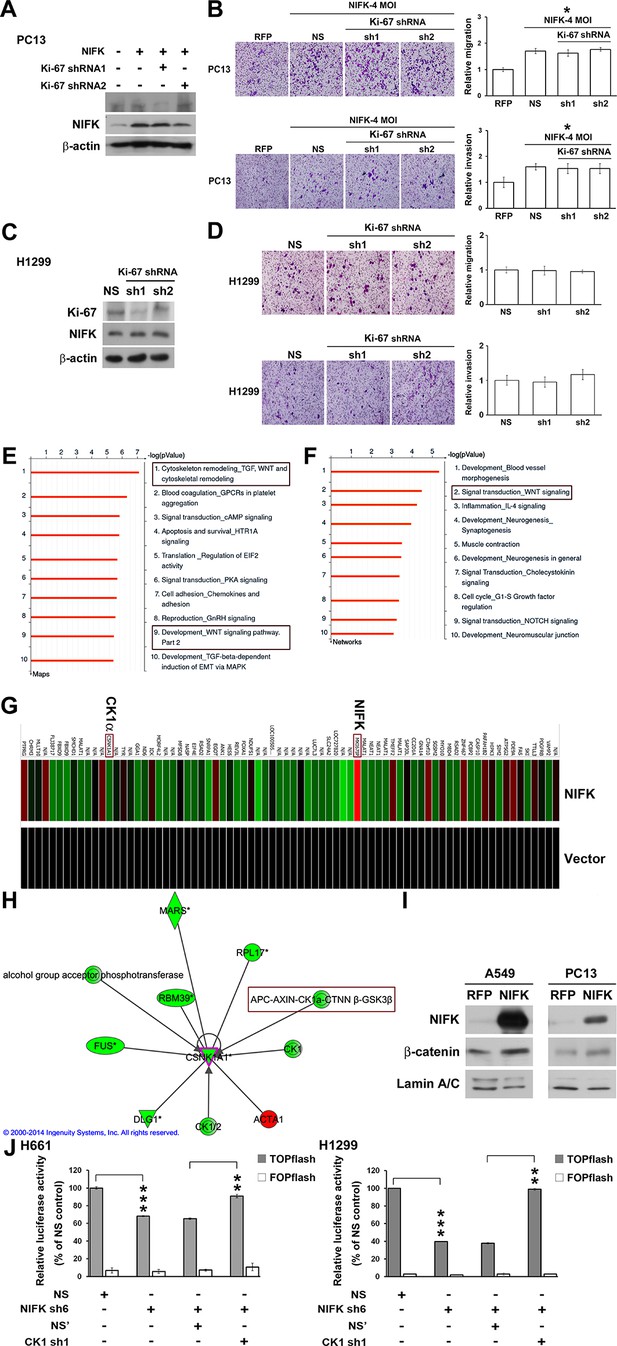
Knowledge-based analysis of the microarray data reveals that NIFK regulate CK1α and Wnt signaling.
(A) The relative knockdown efficiencies of Ki-67 in (A) PC13 and (C) H1299 cells. Cell migration and invasion were evaluated via the transwell assay using (B) PC13 and (D) H1299 cells. (E) List of the top 10 signaling pathways altered by NIFK overexpression. The microarray data of NIFK overexpression in PC13 cells were analyzed using the MetaCore Maps database. (F) List of the top 10 networks affected by NIFK overexpression. The gene signatures were processed via the MetaCore Networks database. (G) Representative signature of genes displaying a≥1.5-fold change in expression due to NIFK overexpression in PC13 cells. The red and green bars represent upregulation and downregulation, respectively. (H) Knowledge-based IPA analysis of the microarray data focusing on CK1α (CSNK1A1)-mediated signaling. The red and green circles represent upregulation and downregulation, respectively. (I) The nuclear levels of β-catenin in cells with NIFK overexpression were showed. Nuclear fractions were extracted from A549 and PC13 cells. (J) TCF/LEF transcriptional activity alteration in the indicated H661 and H1299 cells. TOPflash: reporter plasmid with TCF binding sites. FOPflash: reporter plasmid with mutated TCF binding sites.

NIFK down-regulates CK1α, and up-regulates β-catenin level especially in nucleus.
RT-PCR was performed on RNA samples from PC13 cells overexpressing NIFK.
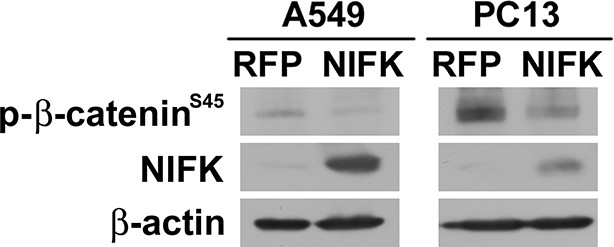
Phospho-β-catenin levels are decreased after NIFK overexpression.
β-catenin phosphorylation was studied in A549 and PC13 cells. Cells were pre-treated with 20 μM proteasome inhibitor MG132 for 3 hr prior lysate harvest for Western blot.
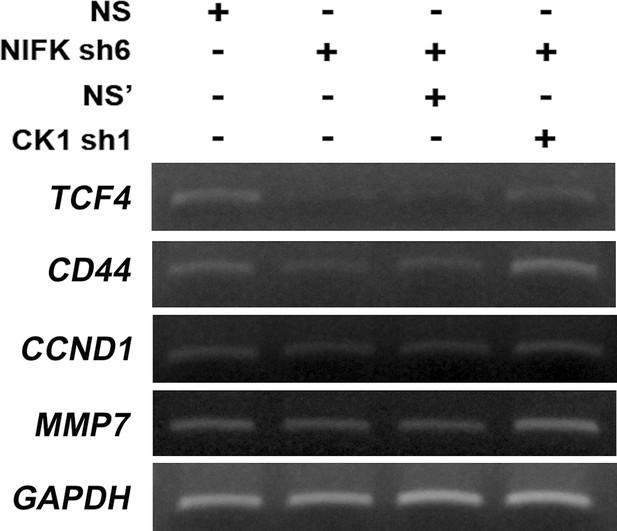
NIFK regulates downstream transcriptional targets of TCF4/LEF via CK1α.
RT-PCR was performed on the indicated samples. The expression levels of transcriptional targets of TCF4/LEF are presented.
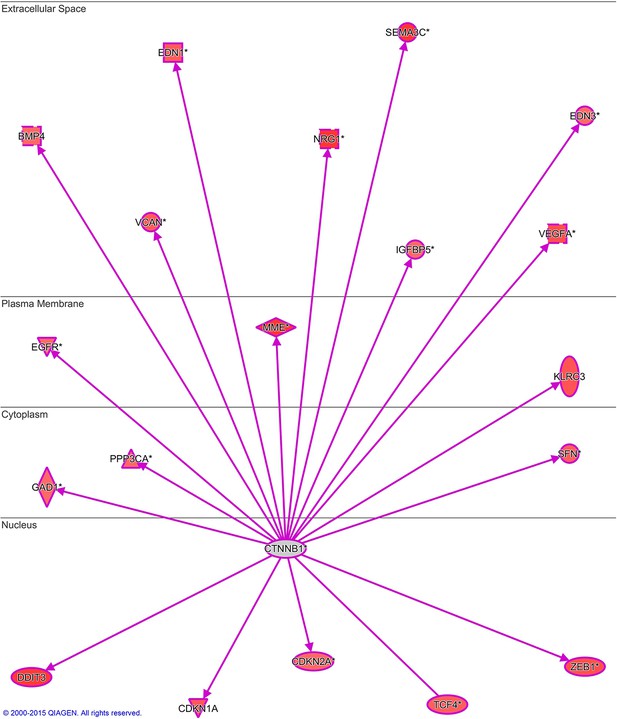
NIFK regulates downstream transcriptional targets of β-catenin.
Microarray analysis of PC13 cells stably overexpressing NIFK by lentiviral infection was performed. β-catenin transcriptional targets upregulated by NIFK were examined and identified via IPA analysis.
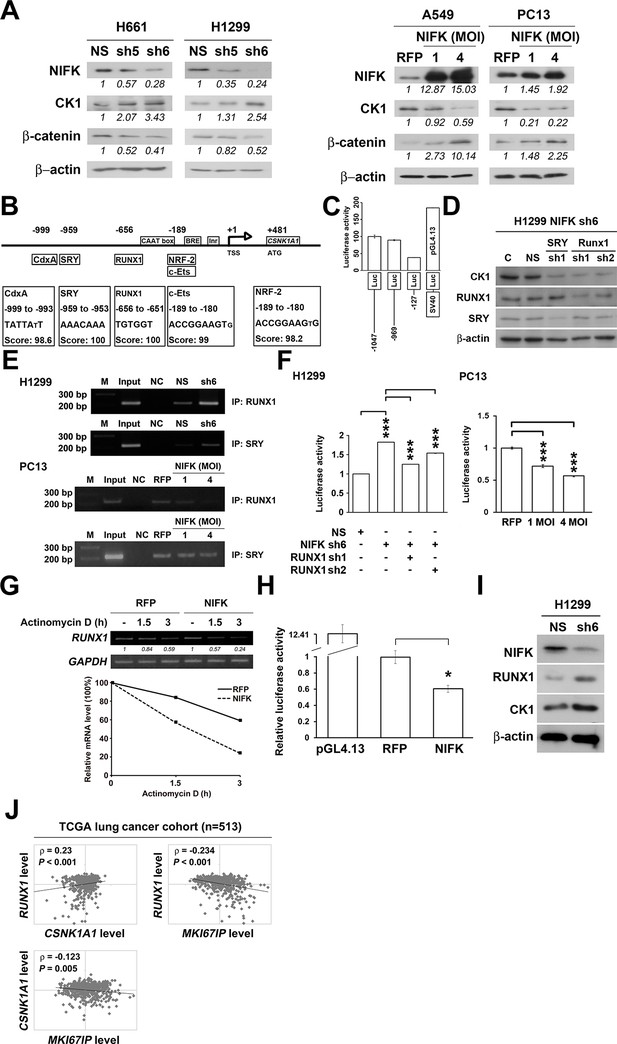
NIFK regulates CK1α expression via the destabilization of transcription factor RUNX1.
(A) Left, the levels of CK1α and β-catenin in H661 and H1299 cells after NIFK knockdown. Right, the levels of the indicated molecules in A549 and PC13 cells upon NIFK overexpression. (B) Identification of cis-regulatory elements within the CSNK1A1 (Ensembl:ENSG00000113712) promoter region. The locations of the consensus binding sites relative to the transcription start site (TSS) are presented below the indicated transcription factors. The scores were calculated by the TFSEARCH website according to the matched sequences. BRE, B recognition element. Inr, initiator element. (C) The CK1 promoter region from -969 to -127 bp is important for transcriptional activation. The relative luciferase activity was measured 48 hr post-transfection with a reporter plasmid containing the CK1 promoter or the indicated deletion mutant. (D) The expression of the indicated molecules after the knockdown of the transcription factors SRY and RUNX1 in the NIFK-silenced H1299 cell clone sh6. (E) ChIP was performed on H1299 (Top) and PC13 cells (Bottom) using antibodies against RUNX1 and SRY. M, DNA marker. NC, negative beads control. (F) A CK1 promoter reporter assay was performed on NIFK-silenced H1299 cells (Left) and in NIFK-overexpressing PC13 cells (Right). The cells were lysed 48 hr after reporter plasmid transfection. The relative luciferase activity was normalized to the number of cells and was quantified. (G) A549 cells were transiently transfected with RFP control or NIFK by lipofection. After 48 h, cells were treated with actinomycin D (5 μg/ml) for indicated time points. Relative RUNX1 mRNA levels were analyzed by RT-PCR. (H) RUNX1 promoter activity was measured after 48 hr of transfection by liposome. (I) NIFK, CK1α and RUNX1 protein levels were analyzed in H1299 cells upon NIFK silencing. (J) Correlation between the expression levels of the indicated molecules in the lung cancer cohort. The microarray data were retrieved from the TCGA database (genomic_TCGA_LUAD_exp_HiSeqV2_percentile_clinical).
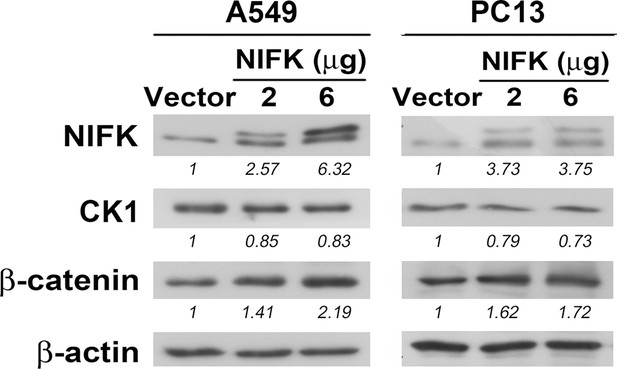
A549 and PC13 cells were transfected with Flag-tagged NIFK or a vector control for 48 hr.
The expression levels of the indicated molecules are presented.
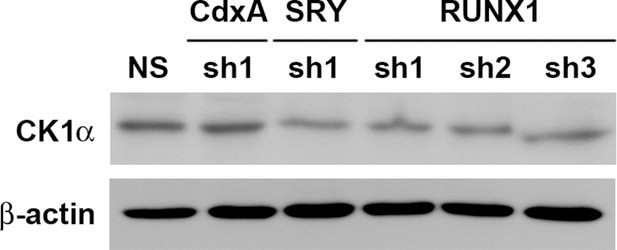
RUNX1 and SRY are potential transcription factors of CK1α.
The expression of CK1α after the knockdown of the candidate transcription factors CdxA, SRY, and RUNX1 in NIFK-silenced H1299 cell clone sh6.
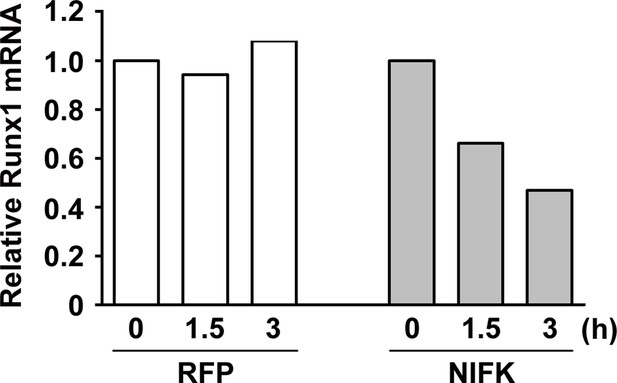
NIFK overexpression leads to RUNX1 mRNA instability.
A549 cells were transfected with RFP control or NIFK by lipofection. Cells were treated with actinomycin D (5 μg/ml) for indicated time points. Relative RUNX1 mRNA levels were analyzed by Q-PCR.
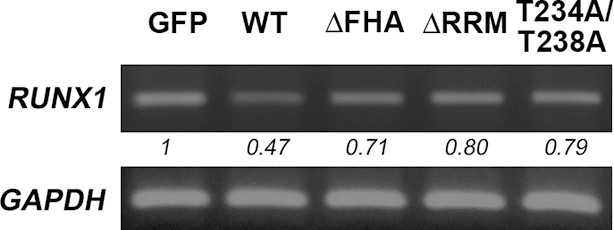
NIFK decreases RUNX1 at RNA level.
Overexpression of GFP-tagged wild type (WT), FHA and RRM truncated and T234A/T238A point mutated NIFK in PC13 cells were performed via liposome and selected. RUNX1 mRNA levels were analysed by RT-PCR.
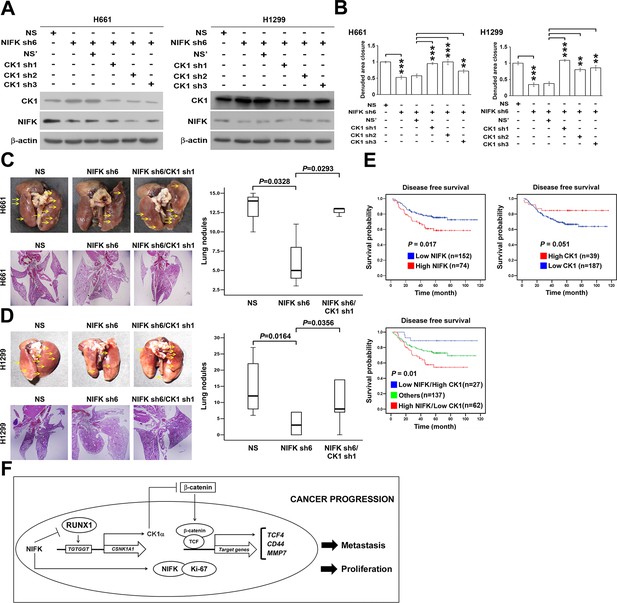
NIFK promotes lung cancer metastasis via CK1α and lung cancer patients with high NIFK/low CK1α represent poor survival rate.
(A) The CK1α knockdown efficiencies of 3 lentivirus-based shRNAs in H661 and H1299 cells. (B) Wound-healing assays were performed on the cells in the indicated groups. The quantification of the migration of H661 and H1299 cells is presented. (C&D) Animal studies were performed on the indicated groups. Left, representative images of lung metastasis and lung HE staining of NSG mice injected with (C) H661 cells or (D) H1299 cells. Right, statistical quantification of the number of metastatic nodules in each group is presented. (E) Kaplan-Meier plot demonstrating the disease-free survival of 226 lung cancer patients displaying varying (upper left) NIFK (MKI67IP) and (upper right) CK1α (CSNK1A1) expression levels. (Lower) Kaplan-Meier plot demonstrating the disease-free survival of cases separated into high NIFK/low CK1α and low NIFK/high CK1α groups. The data were retrieved from the microarray analysis of the GSE31210 dataset. (F) The model of NIFK-induced activation of TCF/β-catenin transcriptional activity via the Runx-1-dependent downregulation of CK1α in metastasis.
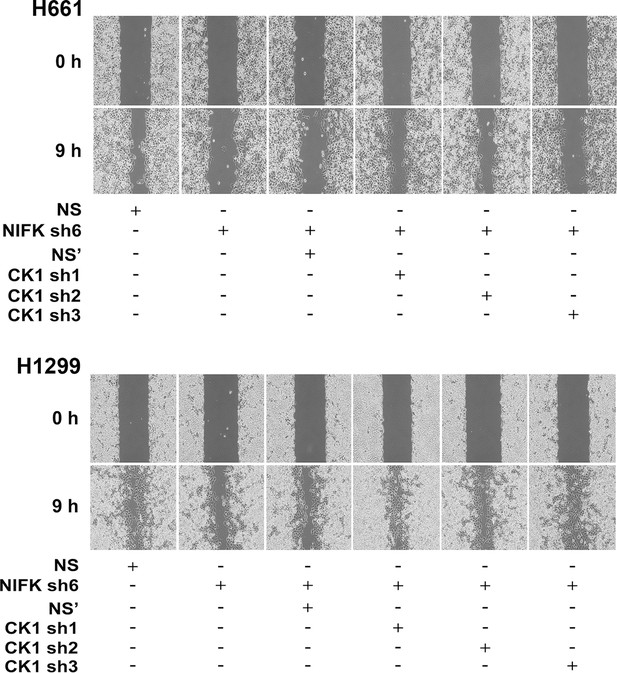
NIFK promotes cancer cell migration via CK1α.
Wound-healing assays were performed on H661 and H1299 cells. Representative images of cell migration in the indicated groups are presented.
Tables
Cox univariate and multivariate regression analysis of CSNK1A1 (CK1α) expression for relapse-free survival in GSE31210 lung cancer dataset
| Univariate | Multivariate | ||||
|---|---|---|---|---|---|
| Survival | Variable | HR (95% CI) | P | HR (95% CI) | P |
| Relapse free (n=246) | CSNK1A1 (high/low) | 2.261 (0.975-5.245) | 0.057 | 1.804 (0.769-4.231) | 0.175 |
| Age | 1.658 (0.984-2.796) | 0.058 | 1.638 (0.97-2.766) | 0.065 | |
| Sex (female/male) | 1.271 (0.778-2.075) | 0.338 | 0.986 (0.489-1.986) | 0.968 | |
| Smoking habit (never/ever) | 1.333 (0.815-2.178) | 0.252 | 1.19 (0.589-2.402) | 0.628 | |
| Stage (I/II) | 3.163 (1.92-5.21) | 0.001 | 2.912 (1.745-4.862) | 0.001 | |
Additional files
-
Supplementary file 1
(A) Gene signatures regulated by lipofection-based NIFK overexpression in PC13 cell were listed.
(B) Signature alternations after lentiviral-based NIFK overexpression in PC13 cell were listed.
- https://doi.org/10.7554/eLife.11288.023





