α/β coiled coils
Figures
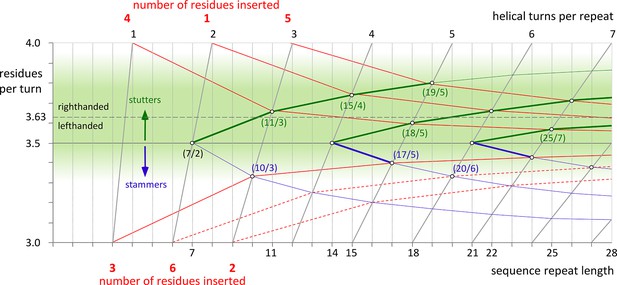
Transitions in periodicity caused by insertions of one to six residues into the heptad repeat.
The green area marks the estimated boundaries of periodicities accessible to α-helical coiled coils. It is centered around the periodicity of unperturbed α-helices, about 3.63 residues per turn. Higher values than 3.63 lead to right-handed and lower values to left-handed supercoiling. The effects of consecutive insertions of stammers (3 residues) or stutters (4 residues) into a heptad pattern are shown by blue and green lines, respectively. The red lines correspond to the insertion of 1 to 6 residues into the heptad periodicity and their progressive delocalization over neighboring heptads. For example, an insertion of 4 residues is accommodated as 11 residues over 3 turns (11/3), when delocalized over one heptad, or as 18/5, when delocalized over two. Insertions of 1 or 5 residues have to be delocalized over two heptads, resulting in periodicities of 15/4 or 19/5 (which could also be brought about by consecutive stutters – following the green line from 7/2 over 11/3 over 15/4 to 19/5). Insertions of 3 can be accommodated as 10/3, at the very edge of the green area, although in the known examples the α-helices are distorted due to the strong left-handed supercoiling which could be avoided by further delocalization. For insertions of 2 or 6 residues (dashed lines) a strong delocalization would be required to reach the green lawn of accessible periodicities. However, for all constructs in this paper, this is not observed. Via the formation of β-layers these insertions sustain the heptad periodicity as unperturbed as possible.
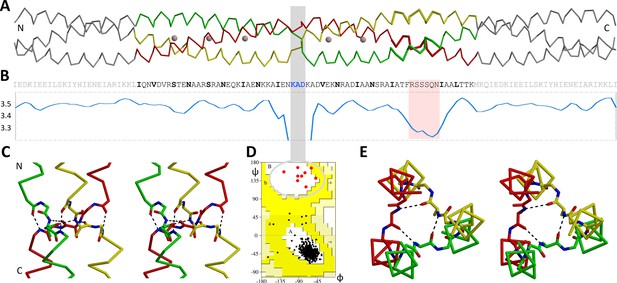
The β-layer in the Actionobacillus OMP100 stalk.
(A) The structure of the Actinobacillus OMP100 stalk construct is aligned with (B) its sequence and a periodicity plot. The area of the stammer is highlighted in pink, the three residues of the β-layer by a grey bar. This bar points to the β region of the Ramachandran plot (D), where all nine β-layer residues of the trimer are found. The close-ups show the (C) side and (E) top view in stereo, highlighting the β-layer interactions. The trimer is colored by chain, GCN4 adaptors in grey. The plot is smoothed over a window of three residues to mask local fluctuations. Empty regions of the Ramachandran plot are cropped.
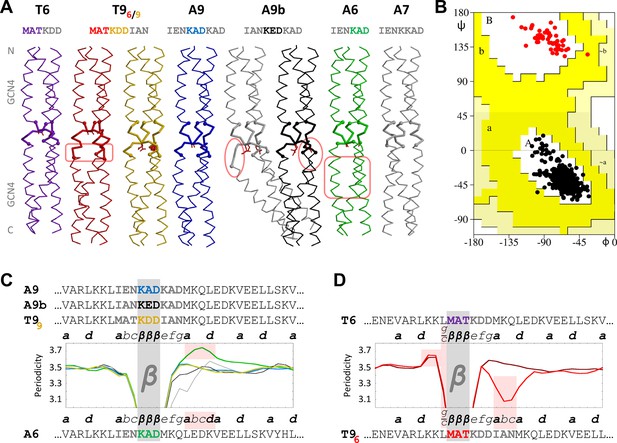
β-layers in 6- and 9-residue motifs between GCN4 adaptors.
(A) The sequences and structures of the GCN4-fusion constructs are shown together with (B) a Ramachandran plot of their backbone torsion angles and (C, D) their periodicities. In the structures, the inserts between the GCN4 adaptors are drawn with thick lines. Disturbances in the α-helical segments are highlighted in pink; the stutter in the A6 structure and the stammer in the T96 structure are also highlighted in pink in panels C and D. In the periodicity plots, all proteins are aligned on the β-layer and their coiled-coil registers are indicated. The plots are shown separately for β-layers forming nonads (C) and hexads (D). A glitch in the periodicity caused by the g/c position preceding β-layers in hexads is highlighted in pink in panel D. As in the previous figure, the periodicity plots are smoothed over a three-residue sliding window. The Ramachandran plot in panel B includes all structures except the kinked grey A9b structure; all residues of the β-layers are shown as red dots and all other residues as black dots. Again, empty regions of the Ramachandran plot are cropped.
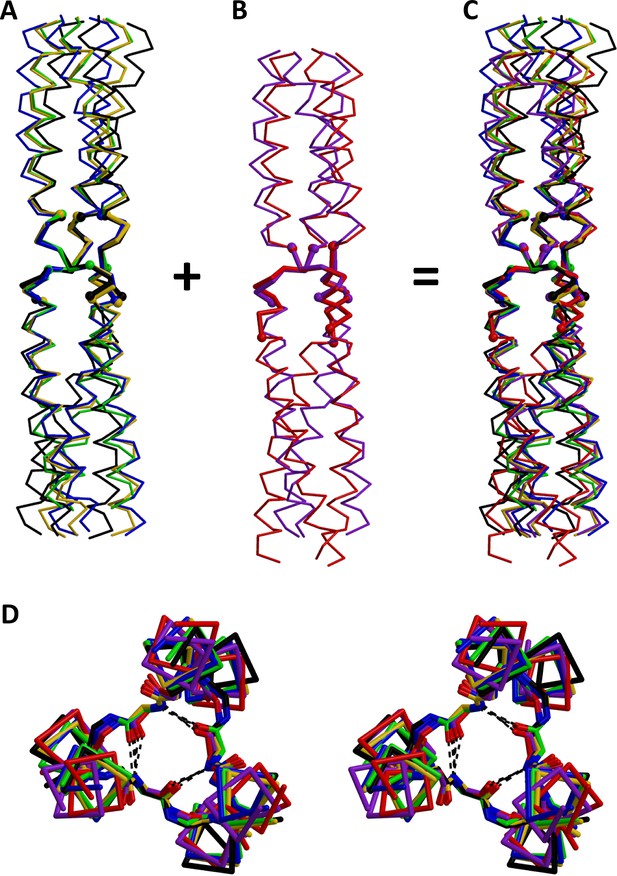
Superimposition of β-layers.
All structures of β-layers between GCN4 adaptors were superimposed on the actual β-layer elements. Superimpositions are shown separately for β-layers occurring (A) in nonads (T99, A9, A9b, A6) and (B) in hexads (T6, T96); the kinked A9b structure (grey in Figure 3) is omitted. Panel (C) shows all β-layers together. (D) Stereo view of the β-layer region in panel C, seen from the N-terminus. The structures are colored as in Figure 3.
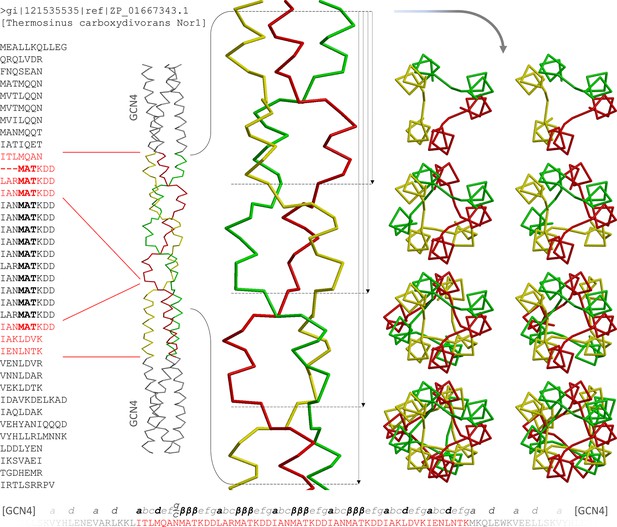
The α/β coiled coil in the Tcar0761 construct.
The two regions fused between GCN4 adaptors in our construct are shown in red on the full sequence of Tcar0761 (left). Next to the sequence, the structure is depicted as a Cα-trace and the four consecutive β-layers are enlarged. On the right, top views are shown, looking down the bundle from the N-terminus. As indicated by the arrows next to the side view, they show 1, 2, 3 or all 4 β-layers. At the bottom, the sequence of the construct is shown together with the assigned register.
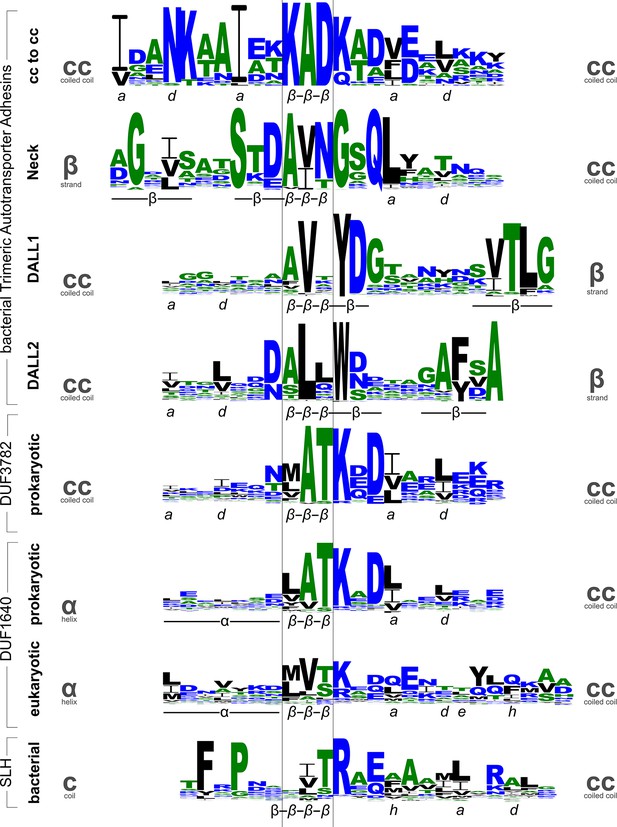
Sequence logos for β-layers in different protein families.
The sequence logos show the conservation patterns of β-layers and their adjacent secondary structure elements in domains of Trimeric Autotransporter Adhesins (stalk, neck, and two variants of the DALL domain), the DUF3782 family of prokaryotic endonucleases, the DUF1640 family of membrane proteins from prokaryotes and organelles, and the surface layer homology (SLH) domain of bacteria. Annotations of the secondary structure (α: helix, β: strand) and coiled-coil register are shown beneath the logos. Grey symbols on the sides indicate the type of secondary structure transition mediated by the β-layer.
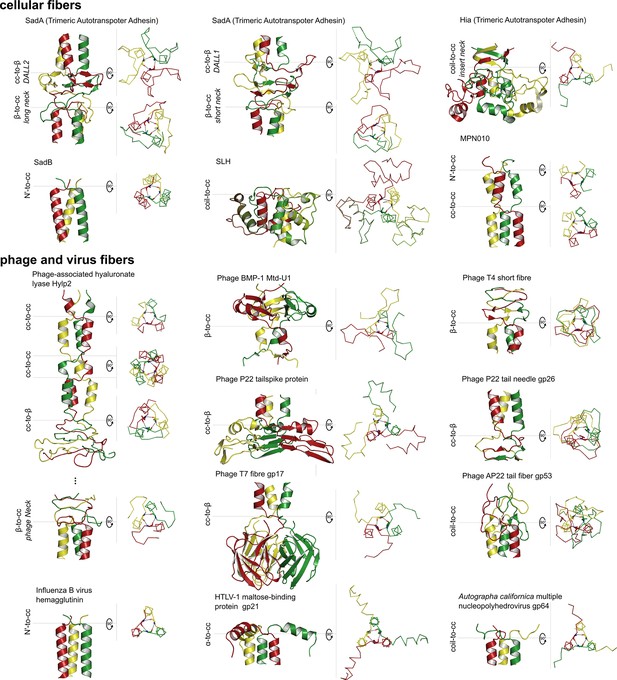
Gallery of canonical β-layers in proteins of known structure.
The parts of the structures containing β-layers are shown in side view (cartoon depiction, left) and the β-layers in top view (backbone trace, right), with their central (β2) residues in stick representation. Table 2 lists the detailed information for the presented proteins.
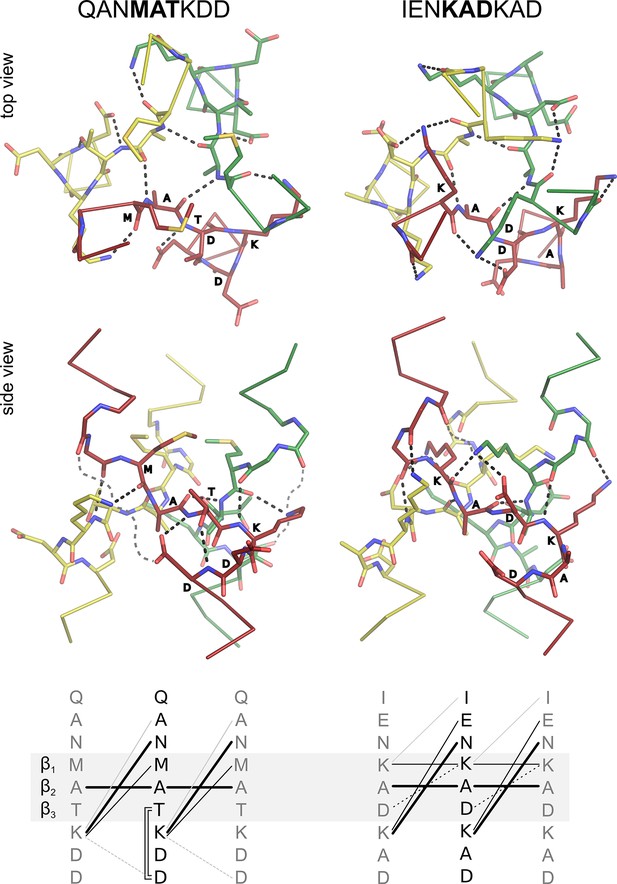
Interaction networks of canonical β-layers.
The two distinct interaction networks of N-capping β-layers based on the sequence MATKDD and C-capping β-layers based on the sequence KADKAD are compared. The upper panels show the interactions at the first β-layer in the Tcar0761 structure and the β-layer in the A9 structure, both in top and side view. For clarity, the side views show only the interactions of the red chain. The lower panels show a schematic representation of the interactions: invariant backbone-to-backbone hydrogen bonds are drawn as bold lines, network-specific backbone-to-sidechain and sidechain-to-sidechain interactions are drawn as solid and broken lines, respectively. Grey lines indicate alternative/additional interactions which are not formed in the depicted β-layers but can be found in other instances as described in the main text. These interactions are indicated by loose grey broken lines in the side views.
Tables
Sequences of constructs and protein buffer composition.
| Construct | Protein sequence | Final buffer |
|---|---|---|
| OMP100 | (GCN4-pII)N-IQNVDVR STENAAR SRANEQK IAENKKA IENKADKAD VEKNRAD IAANSRA IATFRSSSQN IAALTTK-(GCN4pII)c-KLHHHHHH | 20 mM Tris pH 7.5, 400 mM NaCl, 5% Glycerol |
| Tcar0761 | (GCN4-N16V)N-ITLMQAN –––MATKDD LARMATKDD IANMATKDD IANMATKDD IAKLDVK IENLNTK-(GCN4-N16V)c-GSGHHHHHH | 20 mM MOPS pH 7.2, 500 mM NaCl, 5% Glycerol, 2 M Urea |
| T6 | (6xH-TEV)-(GCN4-N16V)N-MATKDD-(GCN4-N16V)c | 20 mM HEPES pH 7.4, 50 mM NaCl, 5% Glycerol, 1 M Urea |
| T9 | (6xH-TEV)-(GCN4-N16V)N-MATKDDIAN-(GCN4-N16V)c | 20 mM HEPES pH 7.4, 50 mM NaCl, 5% Glycerol, 1 M Urea |
| A6 | (6xH-TEV)-(GCN4-N16V)N-IENKAD-(GCN4-N16V)c | 20 mM HEPES pH 7.4, 50 mM NaCl, 5% Glycerol, 1 M Urea |
| A7 | (6xH-TEV)-(GCN4-N16V)N-IENKKAD-(GCN4-N16V)c | 20 mM HEPES pH 7.4, 50 mM NaCl, 5% Glycerol, 1 M Urea |
| A9 | (6xH-TEV)-(GCN4-N16V)N-IENKADKAD-(GCN4-N16V)c | 20 mM HEPES pH 7.4, 50 mM NaCl, 5% Glycerol, 1 M Urea |
| A9b | (6xH-TEV)-(GCN4-N16V)N-IANKEDKAD-(GCN4-N16V)c | 20 mM HEPES pH 7.5, 50 mM NaCl, 10% Glycerol, 1 M Urea |
-
(GCN4-pII)N MKQIEDKIEEILSKIYHIENEIARIKKL
-
(GCN4-pII)C MKQIEDKIEEILSKIYHIENEIARIKKLI
-
(GCN4-N16V)N MKQLEMKVEELLSKVYHLENEVARLKKL
-
(GCN4 N16V)C MKQLEWKVEELLSKVYHLENEVARLKKLV
-
(6xH-TEV) MKHHHHHHPMSDYDIPTTENLYFQGH
β-Layers in proteins of known structure.
| Cellular proteins (canonical) | ||||||
|---|---|---|---|---|---|---|
| PDB | Type | Protein | Domain | Species | Sequence | Similar structures |
| 2YO3 | cc-to-β | SadA | TAA DALL1 | Salmonella enterica | abcdefgβββEEEECC 1306-LKASEAGSVRYETNAD-1321 | 3WPA, 3WPO, 3WPP, 3WQA (Acinetobacter sp. Tol5), 4USX (Burkholderia pseudomallei) |
| 2YO2 | cc-to-β | SadA | TAA DALL2 | Salmonella enterica | abcdefgβββEECCC 310-VAGLAEDALLWDESI-324 | 3ZMF, 2YNZ (Salmonella enterica) |
| 2YO3 | β-to-cc | SadA | TAA Short neck | Salmonella enterica | EEEECCCβββefgabcdefghijklmno 1345-AAVNDTDAVNYAQLKRSVEEANTYTDQK-1372 | 4LGO (Bartonella quintana), 3WP8, 3WPA, 3WPR (Acinetobacter sp. Tol5), 1P9H (Yersinia enterocolitica), 2XQH (Escherichia coli), 3D9X (Bartonella henselae), 2YO0 (Salmonella enterica), 3S6L, 4USX (Burkholderia pseudomallei), 2GR7 (Haemophilus influenzae) |
| 2YO2 | β-to-cc | SadA | TAA Long neck | Salmonella enterica | EEEEβββefgabcdefg 349-DSTDAVNGSQMKQIEDK-365 | 2YNZ, 3ZMF (Salmonella enterica), 3EMO (Haemophilus influenzae), 3LAA, 3LA9, 4USX (Burkholderia pseudomallei), 3WPA, 3WPO, 3WPP, 3WPR, 3WQA (Acinetobacter sp. Tol5), 3NTN, 3PR7 (Moraxella catarrhalis) |
| 1S7M | β-to-cc | Hia | TAA Insert neck 1 | Haemophilus influenzae | EEβββefgabc 642-NTAATVGDLRG-652 | 3EMF (Haemophilus influenzae) |
| 4C47 | Nterm-to-cc | SadB | - | Salmonella enterica | CCβββefgabcdefg 23-DYFADKHLVEEMKEQ-37 | - |
| 5APP | cc-to-cc | OMP100 | TAA Stalk | Actinobacillus actinomycetemcomitans | abcdefgabcβββefgabcdefg 153-IAENKKAIENKADKADVEKNRAD-175 | - |
| 5APZ | cc-to-cc | Tcar0761 | DUF3782 | Thermosinus carboxydivorans | abcdefgβββefgabc 68-ITLMQANMATKDDLAR-83 | - |
| 2BA2 | Nterm-to-cc | MPN010 | DUF16 | Mycoplasma pneumoniae | CCCβββefghijk 5-GTRYVTHKQLDEK-17 | - |
| 2BA2 | cc-to-cc | MPN010 | DUF16 | Mycoplasma pneumoniae | hijkabcβββefgabcdefghijk 14-LDEKLKNFVTKTEFKEFQTVVMES-37 | - |
| 3PYW | coil-to-cc | S-layer protein Sap | SLH | Bacillus anthracis | CCCCEβββefghijkabcdef 35-FEPGKELTRAEAATMMAQILN-55 ... 94-FEPNGKIDRVSMASLLVEAYK-114 ... 156-WEPKKTVTKAEAAQFIAKTDK-176 | - |
| Phage and virus proteins (canonical) | ||||||
| 2C3F | cc-to-cc | Tail fiber hyaluronidase | - | Streptococcus pyogenes (prophage SF370.1) | abcβββefghijkabcβββefgβββefghijk 69-IDGLATKVETAQKLQQKADKETVYTKAESKQE-99 | 2DP5 (Streptococcus pyogenes) |
| 2C3F | cc-to-β | Tail fiber hyaluronidase | - | Streptococcus pyogenes (prophage SF370.1) | defgabcβββCEEEEE 97-SKQELDKKLNLKGGVM-112 | 2DP5 (Streptococcus pyogenes) |
| 2C3F | β-to-cc | Tail fiber hyaluronidase | TAA short neck homolog | Streptococcus pyogenes (prophage SF370.1) | EEEECCEβββefghijkabcdefg 310-DPTANDHAATKAYVDKAISELKKL-327 | 2DP5, 2WH7, 2WB3 (Streptococcus pyogenes) |
| 4MTM | coil-to-cc | gp53 | - | Bacteriophage AP22 | CCCCEβββefgabcdefg 155-NDVGSALSAAQGKVLNDK-172 | - |
| 1YU4 | β-to-cc | Major tropism determinant U1 variant (Mtd-U1) | - | Bordetella Phage BMP-1 | CCCCEEβββefgab 41-TAGGFPLARHDLVK-54 | - |
| 1TSP | cc-to-β | Tailspike protein | Phage P22-tail | Phage P22 | defghijkβββEEE 113-YSIEADKKFKYSVK-126 | 1CLW, 2XC1, 2VFM, 2VFP, 2VFQ, 2VFO, 2VFN [...] (Phage P22) 4OJP, 4OJ5, 4OJL [...] (E. coli Bacteriophage CBA120) 2V5I (Bacteriophage Det7), 2X3H (Enterobacteria phage K1-5) |
| 2POH | cc-to-β | Phage P22 tail needle gp26 | - | Phage P22 | abcdefgβββCEEC 133-ISALQADYVSKTAT-146 | 3C9I, 4LIN, 4ZKP, 4ZKU, 5BU5, 5BU8, 5BVZ (Phage P22) |
| 1H6W | β-to-cc | Short fiber | Receptor binding domain | Bacteriophage T4 | EEEEEECCEEβββefgabcde 321-MTGGYIQGKRVVTQNEIDRTI-341 | 1OCY, 1PDI, 2XGF, 2FKK, 2FL8 (Bacteriophage T4) |
| 4A0T | cc-to-coil | gp17 | gp37_C | Bacteriophage T7 | cdefghijkβββCCCC 454-WLDAYLRDSFVAKSKA-469 | 4A0U (Bacteriophage T7) |
| 1MG1 | α-to-cc | Maltose-binding protein GP21 | TLV_coat | Primate T-lymphotrophic virus 1 (HTLV-1) | HHHHHHEβββefgabcdefghijk 364-AAQTNAAAMSLASGKSLLHEVDKD-387 | - |
| 3DUZ | coil-to-cc | GP64 | Baculo_gp64 | Autographa californica Multiple Nucleopolyhedrovirus | CCCβββefgabcdefg 293-EGDTATKGDLMHIQEE-308 | - |
| 4NKJ | Nterm-to-cc | Hemagglutinin | Hemagglutinin HA2 | Influenza B virus | Eβββefgabcdefghijk 4-VAADLKSTQEAINKITKN-21 | 1QU1 (Influenza A virus) |
| Unusual β-layer proteins | ||||||
| 4NQJ | α-to-α | TRIM Ubiquitin E3 ligase | DUF3583 | Homo sapiens | HHHHHHHβββHHHHHHH 143-SVGQSKEFLQISDAVHF-159 | - |
| 2F0C | (cc-to-)coil-to-β | Receptor binding protein (ORF49) | - | Lactophage tp901-1 | abcdefgabCCCCβββCEEC 22-LEAINSELTSGGNVVHKTGD-41 | 3D8M, 3DA0 (Lactophage tp901-1) |
| 1AA0 | (cc-to-)coil-to-β | Fibritin | Fibritin_C | Bacteriophage T4 | abcdefgCβββEEEEE 450-VQALQEAGYIPEAPRD-465 | 1AVY, 2BSG, 2IBL, 2WW6, 2WW7, 3ALM (Bacteriophage T4), 5C0R (Influenza A), 2LP7 (Human Immunodeficiency Virus 1), 1NAY |
| 2XGF | coil-to-coil | Long tail fiber needle | - | Bacteriophage T4 | EEEECCCCCCCCβββCCCCEEEE 934-EAWNGTGVGGNKMSSYAISYRAG-956 | - |
| 1H6W | coil-to-coil-(to-β) | Short fiber | - | Bacteriophage T4 | CCCβββCCCCEEEEE 284-NADVIHQRGGQTING-298 | - |
| 4UXG | β-to-coil | Proximal long tail fibre protein gp34 | - | Bacteriophage T4 | EEEβββCCCCCC 1233-FVQVFDGGNPPQ-1244 | - |
| 4UXG | α-to-coil | Proximal long tail fibre protein gp34 | - | Bacteriophage T4 | HHHHCβββCCCEEE 1245-PSDIGALPSDNATM-1258 | - |
| 3QC7 | α-to-coil | Head fiber | - | Bacteriophage Phi29 | HHHHHHHβββCCCCCCC 221-NLRTMIGAGVPYSLPAA-237 | - |
Primers used in this study.
| Construct | Primer |
|---|---|
| OMP100 | P omp1: 5`-GACCATGGTCTCCGATTCAGAACGTGGATGTGCGCAGCACCGAAAACGCGGCGCGCAGCCGCGCGAACGAACAG P omp2: 5`-GCTTTATCCGCTTTGTTTTCAATCGCTTTTTTGTTTTCCGCAATTTTCTGTTCGTTCGCGCGGCTGC P omp3: 5`-GAAAACAAAGCGGATAAAGCGGATGTGGAAAAAAACCGCGCGGATATTGCGGCGAACAGCCGCGCGATTGCGACCTTTCG P omp4: 5`-GACCATGGTCTCCTCATTTTGGTGGTCAGCGCCGCAATGTTCTGGCTGCTGCTGCGAAAGGTCGCAATCGCGCG |
| pASK IBA GCN4 N16V | P iba1: 5`-ACAAAAATCTAGATAACGAGGGCAAAAAATGAAACAGCTGGAAATGAAAGTTGAAGAACTGCTGTCCAAAGTCTACCACCTGGAAAACGA P iba2: 5`-CTCGAGGGATCCCCGGGTACCGAGCTCGAATTCGGGACCATGGTCTCCCAGTTTTTTCAGACGCGCAACTTCGTTTTCCAGGTGGTAGAC P iba3: 5`-GTACCCGGGGATCCCTCGAGAGGGGGACCATGGTCTCAATGAAACAGCTGGAATGGAAAGTTGAAGAACTGCTGTCCAAAGTCTACCACC P iba4: 5`-CACAGGTCAAGCTTATTAGTGATGGTGATGGTGATGGCCAGAACCAACCAGTTTTTTCAGACGCGCAACTTCGTTTTCCAGGTGGTAGACTTTGGACAGC |
| T6 | T6 p1: 5`-GGAATTCCATATGAAGCAGCTGGAAGACAAGGTGGAGGAACTGTGTCCAAAGTGTACCATCTGGAAAACGAGGTGGCGCGTCTGAAGAAG T6 p2: 5`-CTTGGACAGCAGTTCTTCCACCTTATCTTCCAGCTGCTTCAATCATCTTTGGTCGCCATCAGCTTCTTCAGACGCGCCACCTC T6 p3: 5`-GGTGGAAGAACTGCTGTCCAAGGTGTATCATCTGGAGAATGAGTGGCGCGTCTGAAGAAGCTGGTGGGCGAACGCTGAGGATCCCG T6 p4: 5`-CGGGATCCTCAGCGTTCGCCCACCAGCTTCTTCAGACGCGCCACTCATTCTCCAGATGATACACCTTGGACAGCAGTTCTTCCACC |
| T9 | T9 p1: 5`-GGAATTCCATATGAAGCAGCTGGAAGATAAGGTGGAAGAGCTGCTGTCAAAGTGTACCATCTGGAAAACGAAGTGGCGCGTCTGAAGAAG T9 p2: 5`-CAGCAGTTCTTCCACCTTATCTTCCAGCTGCTTCATGTTCGCAATGTCATCTTTGGTCGCCATCAGCTTCTTCAGACGCGCCACTTC T9 p3: 5`-GATAAGGTGGAAGAACTGCTGTCCAAAGTGTACCATCTGGAAAACGAAGTGGCGCGTCTGAAGAAACTGGTGGGCGAACGCTGAGGATCCCG T9 p4: 5`-CGGGATCCTCAGCGTTCGCCCACCAGTTTCTTCAGACGCGCCACTTCGTTTTCCAGATGGTACACTTTGGACAGCAGTTCTTCCACCTTATC |
| A6 | A6 p1: 5`-GGAATTCCATATGAAGCAACTTGAAGACAAAGTCGAAGAGCTTCTCTCAAGTTTATCATCTTGAGAACGAAGTTGCTCGTCTTAAG A6 p2: 5`-CCTTAGAAAGAAGTTCTTCGACCTTATCCTCAAGTTGCTTCATATCGCTTTGTCTCAATGAGTTTCTTAAGACGAGCAACTTCG A6 p3: 5`-CGAAGAACTTCTTTCTAAGGTTTACCATCTCGAAAATGAGGTTGTCGTTCAGAAGCTTGTTGGCGAACGCTGAGGATCCCG A6 p4: 5`-CGGGATCCTCAGCGTTCGCCAACAAGCTTCTTGAGACGAGCAACCCATTTCGAGATGGTAAACCTTAGAAAGAAGTTCTTCG |
| A7 | MP A6+K se: 5`-CTTAAGAAACTCATTGAGAACAAGAAAGCCGATATGAAGCAAC MP A6+K as: 5`-GTTGCTTCATATCGGCTTTCTTGTTCTCAATGAGTTTCTTAAG |
| A9 | MP A6+KAD se: 5`-CATTGAGAACAAAGCCGATAAGGCTGACATGAAGCAACTTGAGG MP A6+KAD as: 5`-CCTCAAGTTGCTTCATGTCAGCCTTATCGGCTTTGTTCTCAATG |
Crystallization and cryo condition.
| Structure | Protein solution & concentration | Reservoir solution (RS) | Cryo solution |
|---|---|---|---|
| OMP 100 | 20 mM Tris pH 7.5, 150 mM NaCl, 3% (v/v) Glycerol, 3 mg/ml protein | 0.1 M tri-Sodium citrate pH 5.5, 2% (v/v) Dioxane 15% (w/v) PEG 10,000 | RS + 15% (v/v) PEG 400 |
| A6 | 20 mM HEPES pH 7.2, 50 mM NaCl, 2% (v/v) Glycerol, 1 M Urea, 15 mg/ml protein | 95 mM tri-Sodium citrate pH 5.6, 19% (v/v) Isopropanol, 19% (w/v) PEG 4000, 5% (v/v) Glycerol | - |
| A7 | 20 mM HEPES pH 7.3, 50 mM NaCl, 1 M Urea, 15 mg/ml protein | 0.1 M Citric acid pH 3.5, 3 M NaCl | - |
| A9 | 20 mM HEPES pH 7.2, 50 mM NaCl, 2% (v/v) Glycerol, 1,5 M Urea, 17 mg/ml protein | 1.6 M tri-Sodium citrate pH 6.5 | - |
| A9b black | 50 mM HEPES, 50 mM NaCl, 1 M Urea, 7.5 mg/ml protein | 2.4 M Sodium malonate pH 5.0 | - |
| A9b grey | 50 mM HEPES, 50 mM NaCl, 1 M Urea, 7.5 mg/ml protein | 0.2 M Sodium citrate, 0.1 M Bis Tris propane pH 6.5, 20% (w/v) PEG 3350 | - |
| T6 | 20 mM HEPES pH 7.2, 50 mM NaCl, 1 M Urea, 13 mg/ml protein | 0.2 M CaCl2, 0.1 M HEPES pH 7.5, 30% (w/v) PEG 4000 | - |
| T96 | 20 mM HEPES pH 7.2, 50 mM NaCl, 2% (v/v) Glycerol, 1.5 M Urea, 15 mg/ml protein | 0.2 M Ammonium phosphate, 0.1 M TRIS pH 8.5, 50% (v/v) MPD | - |
| T99 | „ | 0.1 M Citric acid pH 5.0, 20% (v/v) Isopropanol | RS + 1 M Urea +25% Glycerol |
| Tcar 0761 | 20 mM MOPS pH 7.2, 400 mM NaCl, 5% (v/v) Glycerol, 1.5 M Urea, 7 mg/ml protein | 0.1 M tri-Sodium citrate pH 4.0, 30% (v/v) MPD | - |
Data collection and refinement statistics.
| Structure | OMP100 | A6 | A7 | A9 | A9b black | A9b grey | T6 | T96 | T99 | Tcar0761 |
|---|---|---|---|---|---|---|---|---|---|---|
| Beamline/Detector* | PXII / M | PXII / M | PXII / M | PXIII / M | PXII / P | PXII / P | PXII / P | PXII / M | PXIII / M | PXII / P |
| Wavelength (Å) | 0.9786 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| Trimers/AU | 1 | 1 | 1/3 | 1 | 1 | 2 | 1 | 1 | 1 | 1/3 |
| Space group | C2** | C2** | P321 | P21 | P21 | P21 | P21 | P21 | C2** | P63 |
| a (Å) | 62.1 | 60.4 | 38.2 | 65.2 | 26.2 | 71.1 | 34.2 | 25.1 | 60.8 | 37.9 |
| b (Å) | 35.9 | 34.8 | 38.2 | 34.6 | 37.5 | 35.0 | 27.0 | 38.3 | 35.1 | 37.9 |
| c (Å) | 198.5 | 104.2 | 87.1 | 67.5 | 95.0 | 106.2 | 101.0 | 105.0 | 112.2 | 179.2 |
| β (°) | 96.0 | 101.1 | 90 | 117.7 | 92.6 | 101.7 | 93.9 | 93.3 | 100.4 | 90 |
| Resolution range (Å)*** | 32.9–2.30 (2.44–2.30) | 30.0–2.10 (2.23–2.10) | 18.2–1.37 (1.45–1.37) | 33.7–1.80 (1.91–1.80) | 34.9–1.35 (1.43–1.35) | 38.1–2.00 (2.12–2.00) | 34.1–1.60 (1.70–1.60) | 34.9–1.80 (1.91–1.80) | 19.5–2.00 (2.12–2.00) | 32.3–1.60 (1.69–1.60) |
| Completeness (%) | 92.4 (86.5) | 97.3 (96.2) | 99.0 (98.6) | 98.9 (97.4) | 95.9 (92.1) | 92.4 (98.9) | 98.2 (96.1) | 97.1 (95.4) | 98.7 (97.5) | 99.2 (96.9) |
| Redundancy | 2.84 (2.52) | 3.71 (3.71) | 6.35 (6.33) | 3.70 (3.67) | 3.72 (3.47) | 3.29 (3.31) | 3.04 (2.89) | 3.94 (3.81) | 3.73 (3.73) | 3.69 (3.65) |
| I/σ(I) | 14.0 (1.88) | 15.5 (2.28) | 18.2 (2.52) | 14.3 (2.07) | 17.6 (2.10) | 13.9 (2.43) | 13.6 (2.33) | 14.5 (2.14) | 19.5 (2.25) | 20.3 (2.23) |
| Rmerge (%) | 4.2 (44.8) | 4.8 (62.1) | 5.1 (75.5) | 5.1 (61.7) | 3.4 (66.6) | 5.0 (51.5) | 4.4 (42.3) | 7.2 (71.7) | 4.0 (63.2) | 2.9 (60.2) |
| Rcryst (%) | 22.5 | 20.8 | 19.5 | 20.6 | 16.3 | 20.6 | 17.4 | 18.7 | 21.1 | 17.7 |
| Rfree (%) | 25.4 | 25.1 | 23.8 | 25.6 | 19.9 | 25.3 | 20.5 | 22.6 | 25.5 | 21.3 |
| PDB code | 5APP | 5APQ | 5APS | 5APT | 5APU | 5APV | 5APW | 5APX | 5APY | 5APZ |
-
*M = MARRESEARCH mar225 CCD detector; P = DECTRIS PILATUS 6M detector
-
**twinned with apparent H32 symmetry and twinning operators
-
1/2*h-3/2*k,-1/2*h-1/2*k,-1/2*h+1/2*k-l and 1/2*h+3/2*k,1/2*h-1/2*k,-1/2*h-1/2*k-l
-
***values in parenthesis refer to the highest resolution shell





