Brain-wide mapping of neural activity controlling zebrafish exploratory locomotion
Figures
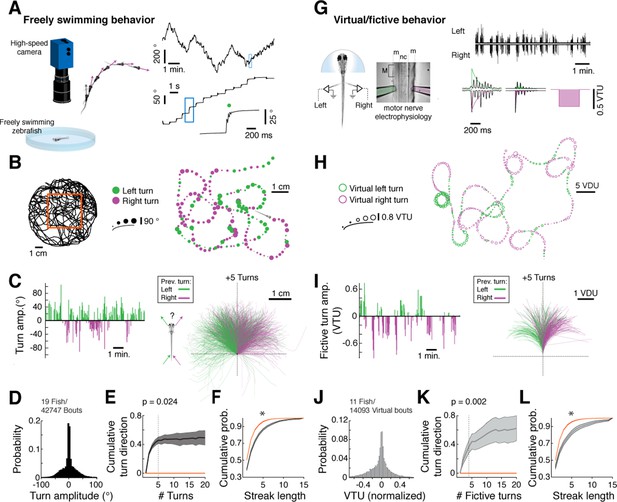
Spontaneous orienting behavior is governed by switches in turn state.
(A) As fish explore a homogeneous environment, heading direction (purple vectors) over time is recorded with a high-speed camera. Fish execute discrete spontaneous turns (top right, showing heading direction over time) that comprise sequences of turns biased in the same direction (middle right). The size and color of the spot (bottom right) denote the magnitude and direction of the underlying turn, respectively. (B) Plot of a swim trajectory taken from a much longer recording (from within red box, left). Turn direction is encoded by color (left turns in green; right turns in magenta). Dots are positioned at the points in the trajectory where turns were executed; dot size is proportional to turn angle. Note the chains of left and right turns that confer a characteristic slaloming shape to the swim trajectory. (C) Left, turn states can also be visualized by plots of turn amplitude over time, colored according to turn direction. Notice that the fish tends to turn in streaks. Right, when swim trajectories are triggered, rotated, aligned, and color-coded according to the direction of each preceding turn, it is again evident that the previous turn biases the future trajectory of the animals in the same direction. (D) Histogram of turn amplitudes from 42,747 swim events across 19 fish. The overall turn distribution is symmetric. (E) Quantification of average turn history-dependence. After a left -> right or right -> left switch event, fish tend to turn with a bias in the same direction for 5 swim bouts (p=0.024 for the change in cumulative turn direction, signed rank test compared to a randomly turning model fish, shown in red). N = 19 fish. Shaded error is SEM across fish. (F) Cumulative histogram of streak length (number of turns in the same direction before a switch) for 19 fish, black, compared to randomly turning fish, red. (*) p<10–5 rank sum test. Shaded error is SEM across fish. (G) Turns can also be decoded from electrophysiological recordings from peripheral motor nerves in paralyzed fish. Turn direction and amplitude are calculated by subtracting the normalized power of recorded bursts in the left channel (green electrode) from bursts in the right channel (pink electrode), weighing the start of a burst more than the end (see exponential filters, right, and 'Materials and methods'). (m) muscle; (nc) notochord; M (myotome); VTU (virtual turn unit). (H) Sequences of decoded virtual turns and virtual swim distances (sum of left and right fictive channels, virtual distance units [VDU]) can be used to plot virtual swim trajectories. The pattern of unidirectional sequences observed in freely swimming fish is conserved in fictively swimming fish. (I) Fictive turn amplitude and trajectory history plot for the data in (G) and (H). (J) Histogram of fictive turn amplitudes from 14,093 swim events in 11 fish. (K) Across animals, fictively swimming fish tend to turn with a bias in the same direction for 4 swim bouts after a change in turn direction (p=0.002 for the change in cumulative turn direction, signed rank test compared to a randomly turning model fish, red), although many chains persist for much longer. N = 11 fish. Shaded error is SEM across fish. (L) The cumulative probability distribution of fictive streak length is also significantly different from a randomly turning model fish. (*) p<10–4, rank sum test. N = 11 fish. Shaded error is SEM across fish.
-
Figure 1—source data 1
Behavioral data from freely swimming larval zebrafish, with analysis code.
This zip file contains the swim data (Matlab -- figure1e_sourcedata.mat) and analysis scripts (Matlab -- figure1e.m, cumulativespont.m) used to generate Figure 1E. Please refer to figure1e.m for specific instructions.
- https://doi.org/10.7554/eLife.12741.004
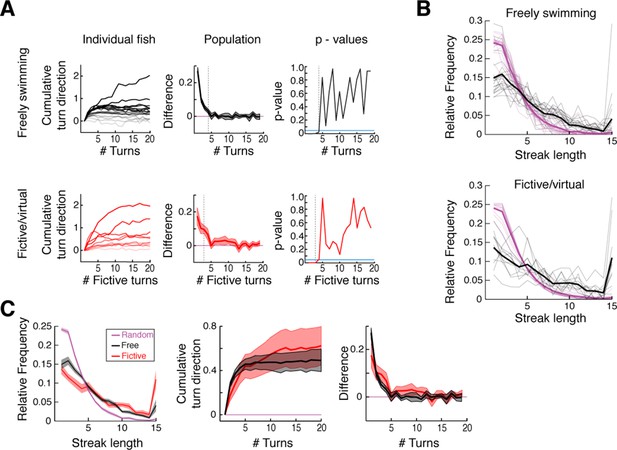
Analysis of free and fictive turn states.
(A) Signed cumulative turn angle triggered on left->right or right->left switch events for individual fish (left column) and change in signed turn angle across fish (middle column) for freely swimming (top row) and fictively swimming (bottom row) fish. A p-value threshold of 0.05, for a signed rank test between the change in cumulative turn angle and a randomly turning fish (blue line, 0) at each turn since a switch event, is used to determine the average length of turn states in Figure 1E,K. (B) Black, histograms of streak length for all freely swimming and fictively swimming fish (top and bottom, respectively). Magenta, corresponding histograms of expected random streak lengths given overall turn biases for each fish. Thick lines are the mean for each data set. Here, relative frequency is defined as the fraction of all bouts spent inside a streak of a given length. (C) Direct comparison of streak statistics for freely and fictively swimming fish from (B). Note the substantial overlap of streak length, cumulative turn direction, and change in turn direction plots. Shaded error is SEM.
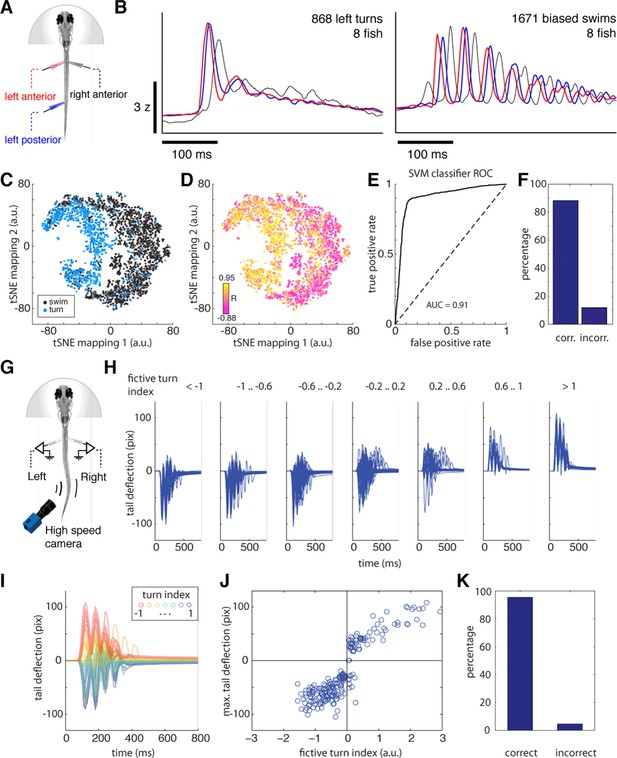
Fictive swimming is a reliable readout of intended locomotion.
(A) Schematic of three-electrode motor nerve recording experiment. Anterior and posterior electrodes were at least 12 body segments apart. Motor nerve volleys in the left and right anterior electrodes were used to classify motor events into left turns, right turns and biased forward swims, as in the data presented in Figures 1 and 2. To verify that these turns were classified correctly, fictive waveforms from the anterior and posterior electrodes were then compared. (B) Left, average fictive waveform recorded on the left anterior (red), left posterior (blue), and right anterior (black) electrodes for 868 fictive bouts classified as left turns from 8 fish (using only the anterior electrode signals for classification). The simultaneous rise time of the first burst on the ipsilateral anterior and posterior electrodes is characteristic of a turning maneuver, as turns are typically initiated with simultaneous contractions of ipsilateral musculature. Right, average fictive waveform recorded on the left anterior (red), left posterior (blue), and right anterior (black) electrodes for 1671 fictive bouts classified as biased forward swims from 8 fish (using only the anterior electrode signals for classification). The phase lag between the first burst on the ipsilateral anterior and posterior electrodes is characteristic of a biased forward swim, in which a near-symmetrical wave propagates down the tail. (C) Analysis of single motor events. Colors represent, for individual motor events, whether it is classified as a turn (blue) or forward swim (black) using only the anterior two electrodes. The 2D space represents a dimensionally reduced representation – using tSNE (t-Distributed Stochastic Neighbor Embedding) – of the two electrodes on the left side of the tail (right side for some fish), ignoring the right (left for some fish) anterior electrode. Thus, although the 2D space is built only from features representing the anterior and posterior left tail signals, swims classified by only the anterior signals segregate, forming evidence that only the anterior signals are sufficient for classifying swim direction. (D) The correlation coefficient (R) between the first bursts on the ipsilateral anterior and posterior electrodes within tSNE space. Although R was not used to build the tSNE space, low and high values can be seen to approximately segregate, showing that the overlap – a proxy for phase – between anterior and posterior signals is represented in the tSNE space. (E) Receiver-operator characteristic (ROC) curve for a Gaussian support vector machine (SVM) classifier trained on the tSNE separation of turns and swims with 5-fold cross-validation (using drtoolbox, http://lvdmaaten.github.io/drtoolbox/ and Classification Learner, Mathworks), which classifies bout type with 88.2% accuracy (F). Some of the errors will be due to noise in the recording or suboptimal classification. This result shows that classifying motor events by the anterior electrode signals yields results consistent with considering three electrode signals, showing that the left and right anterior signals contain accurate information for turn direction. (G) Independent experiment for assessing reliability of using fictive recordings for decoding motor events. Schematic of experiment in which both fictive signals and tail motion are monitored in weakly paralyzed fish. Two fictive electrodes record fictive swim patterns, while small undulations in the weakly paralyzed tail are filmed with a high-speed camera. (H) Filmed tail motion for each bout classified into turns and swims based only on the fictive recordings in an example fish, showing that tail movement waveforms are consistently related to fictive turn index. (I) Partially paralyzed tail waveforms colored according to each associated fictive turn index. For ease of visibility, only the first 10 motor events per turn index range are shown. (J) Scatterplot of fictive turn index and maximum tail deflection. Higher amplitude fictive turns are associated with higher amplitude tail deflections. (K) The turn/swim classification error rate for the weakly paralyzed experiment. Motor events with absolute fictive turn angle less than 0.05 were not considered turns. Only 4% of turns are misclassified, the remaining 96% matching between fictive and physical direction. n = 47,58,72,65,25,12,22 turns.
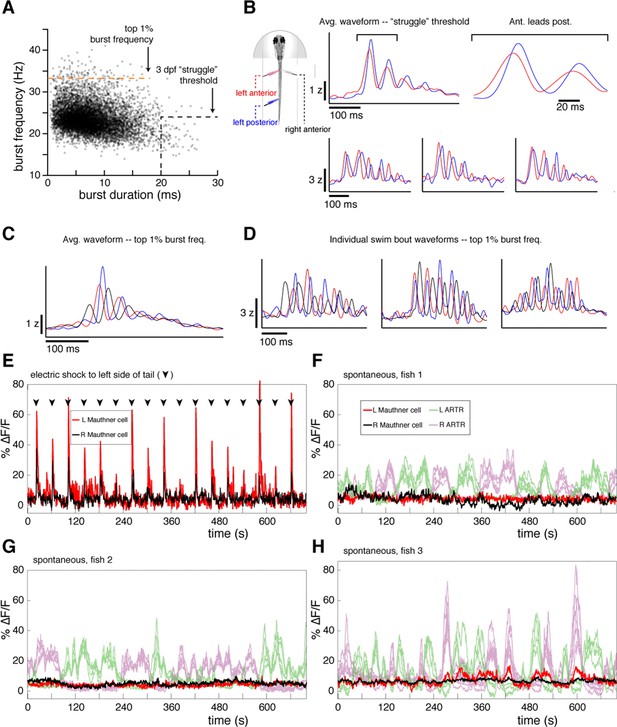
Fictive swims are not struggles or startles.
(A) Scatterplot of burst duration and burst frequency for n = 8466 fictive swim bouts across N = 9 fish that were measured with three recording electrodes. Black dotted line, threshold for fictive struggles as determined in 3 dpf fish in (Liao and Fetcho, 2008). Orange dotted line, top 1% of all burst frequencies. While this percentile falls far short of the burst frequencies expected during escapes (33.5 Hz here, vs. 56.6 Hz in [Budick and O'Malley, 2000], and 62.7 Hz in [Dunn et al., 2016]), we used it to isolate events that were investigated in more detail. (B) Left, schematic of recording conditions. The colors of plots in (B–D) correspond to the color scheme for each electrode. Right, average (top) and example (bottom) waveforms recorded from the ipsilateral anterior and posterior electrodes for bouts that met published 3 dpf struggle criteria. Note that waves propagate rostral-to-caudal as in normal swimming behavior. (C) Average waveforms recorded from all electrodes for bouts in the top 1% of the burst frequency distribution, showing no sign of waves propagating caudal-to-rostral, a hallmark of struggles (Liao and Fetcho, 2008). (D) Three example bouts illustrating that even bouts with fast burst frequencies resemble normal swimming waveforms with no sign of waves traveling caudal-to-rostral. (E) Fluorescence time series from the left and right Mauthner cell (M-cell) in a single Tg(elavl3:H2B-GCaMP6f) larva during electric shock stimulation (arrowheads) (M-cells identified using spinal backfills). Electric shocks tend to elicit escapes with associated M-cell activity; here, the shocks can be seen to evoke M-cell activity consistently. (F–G) Three examples of M-cell activity from three fish under spontaneous conditions (M-cells identified using spinal backfills). While the ARTR is highly active, the M-cells are mostly quiescent. Subthreshold activation can be observed on occasion, but transients are never seen at the magnitude associate with escape behavior during electric shock (E), suggesting that fictive escapes are rare if not altogether absent in our spontaneous fictive swimming experiments.
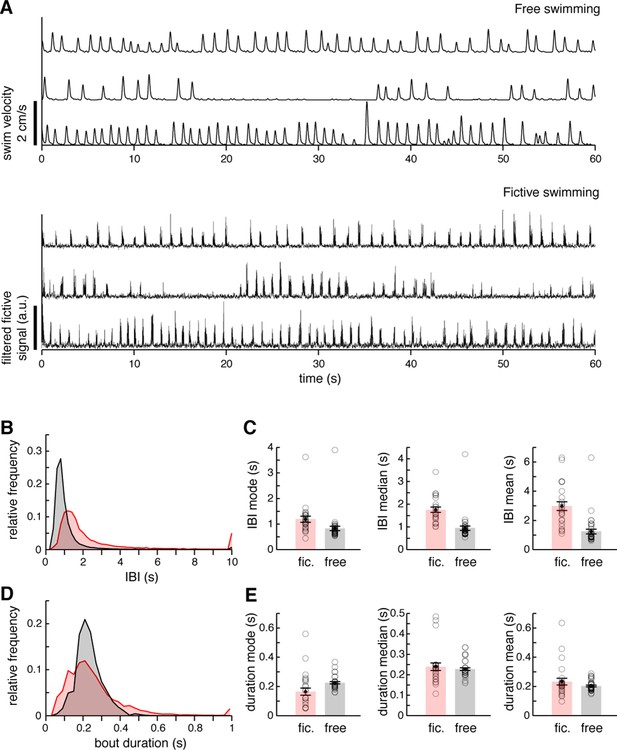
Comparison of free and fictive swimming statistics.
(A) Top, three example records of instantaneous swim velocity for freely swimming fish, illustrating the variability in bout frequency across periods and fish. Velocity is derived from a lightly smoothed record of trajectory (see 'Materials and methods'). Bottom, three example records of filtered fictive amplitude for fictively swimming fish, illustrating the variability in fictive bout frequency across periods and fish. (B) Interbout interval (IBI) histograms for N = 37 freely swimming fish and N = 25 fictively swimming fish. The distributions overlap significantly, but fictively swimming fish are sometimes inactive for longer periods. (C) Quantifications of IBI mode, median, and mean for both free and fictive swimming, averaged across fish. (D) Swim bout duration histograms. The distributions overlap significantly, although fictive durations are more variable. Duration measurements are from the same fish as in (B). (E) Quantifications of bout duration mode, median, and mean for both free and fictive swimming, averaged across fish.
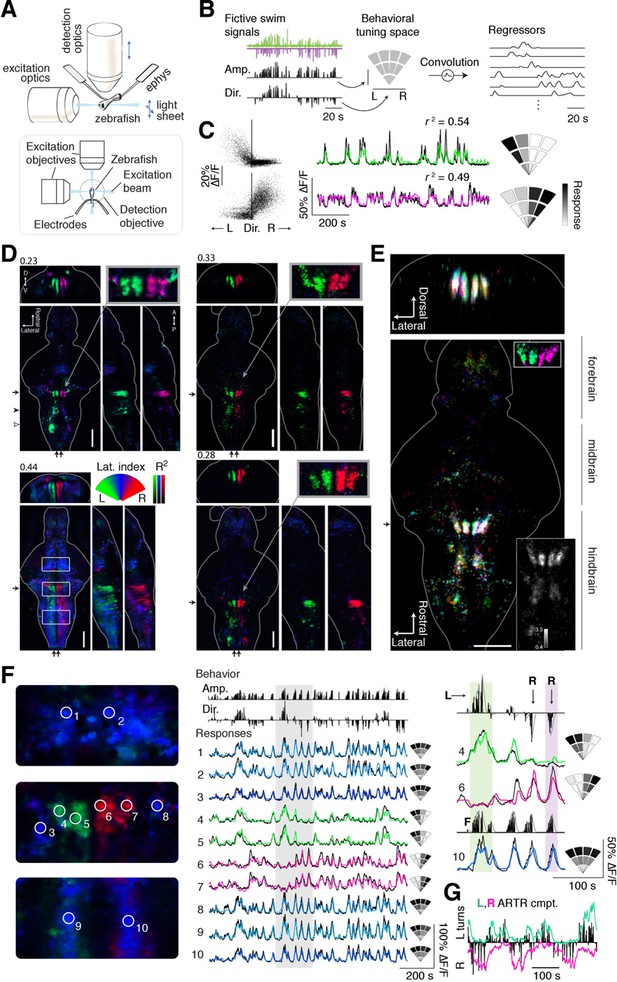
Whole-brain analysis identifies neural structures correlated with turning behavior.
(A) Schematic of experimental paradigm for fictive swimming combined with light-sheet imaging ('Materials and methods'). (B) Schematic of analysis technique. Left: First, fictive swim signals are converted into measures of swim amplitude ('Amp') and turning direction ('Dir' for laterality). Middle: Next, amplitude and laterality are mapped onto the vertical and horizontal axes of a 2D space. This space is tiled with 12 basis functions, each representing a region in this 2D behavior space, now defined in polar coordinates ('Materials and methods'). Contours are shown for clarity; actual basis functions overlap by 50%. Right: The signal from each bin is convolved with an impulse response function to generate a regressor; an example subset of regressors is shown. (C) Brain activity is regressed against the regressors constructed in (B) to generate a behavioral tuning function for every voxel. Voxels of two example neurons are shown here. Left, relationship between turn laterality and neural response for the two example neurons, each dot is a time point. Middle, time series from the same two example neurons. Black line, ∆F/F; colored line, prediction of best-fitting model (see panel B). Right, behavioral tuning for the same two neurons, given by regression coefficients, using the analysis described in panel B; grayscale ranges from 10th to 90th percentile of the coefficient weights. (D) Behavioral tuning maps across the brain derived from fitting every voxel with the regressors described in panel B, for four representative fish. Calcium indicators are either localized in cytoplasm (left two fish) or in the nucleus (right two fish). The dorsal view is a maximum intensity projection over the whole brain; the side and front views are taken from a maximum intensity projection of 21 slices (~10 μm) along the medial-lateral axis and rostral-caudal axis, respectively. Numbers above each panel indicate the R2 value at which the color map saturates (maximum R2 value is higher), color maps start at R2 = 0. Arrows in each panel represent the centroid position of these slices for the frontal view (top) or side view (right). Solid arrowhead: diffusive correlated region in rhombomeres 4–6. Open arrowhead, inferior olive. Scale bar, 100 μm. D, dorsal; V, ventral; A, anterior; P, posterior. (E) Registered map from seven different fish (nuclear localized GCaMP6f) to a standard brain. Each fish is encoded by a different color; brightness represents R2. Bottom, top-down maximum intensity projection (along the dorsal-ventral axis). Top, front projection as in d, with the centroid of the slice indicated by the arrow in bottom panel. Top right inset, ARTR region across fish in the standard brain, but with color representing laterality as in panel d, showing consistent tuning across animals. Bottom right inset, a measure of stereotypy in location of functionally identified neurons across the 7 fish. Intensity represents the standard deviation divided by the mean of R2 (thresholded at 0.04). Scale bar, 100 μm. (F) Example ΔF/F traces from regions of interest (ROIs) in panel (D) (left bottom, white boxes). Left, top to bottom: midbrain, ARTR, and caudal hindbrain. Middle, top, signals of swim amplitude (Amp.) and turn laterality (Dir.). Black bars represent several individual swim events. Bottom, ∆F/F from ROIs in the left panels. Right, enlarged view of gray region in middle panel. L,R,F stand for left turns, right turns and swim amplitude, respectively. Responses from ROIs 1–3 and 8–10 show tuning to swim amplitude; ROIs 4,5 to left turns, and ROIs 6,7 to right turns. (G) In addition to single cells, activity of left and right populations derived with ICA (Figure 2—figure supplement 1C–E; bottom-right fish of Figure 2D) tracks turning behavior.
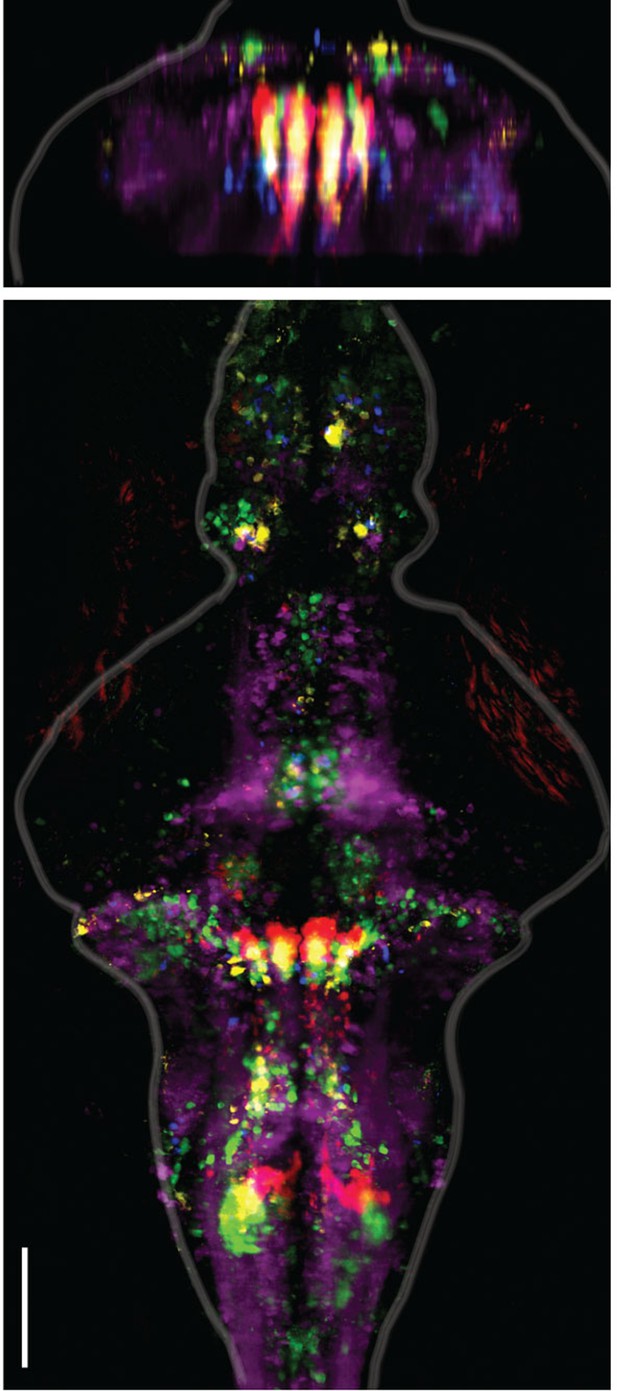
Alignment of functional brain maps in fish expressing calcium indicators in the cytosol.
Shown here is the overlay of brain maps of transgenic zebrafish expressing calcium indicators in the cytosol of neurons (Tg(elavl3:GCaMP6f) and one Tg(elavl3:GCaMP5G) fish). In comparison to Figure 2E, which shows the overlaid maps for fish expressing calcium indicators only in the nuclei of neurons, the functionally identified neurons occupy the same locations, co-localize across fish, and are consistent with the locations of the ARTR as identified in Ahrens et al. 2013. Analyses are identical to those used in Figure 2. Scale bar, 100 μm.
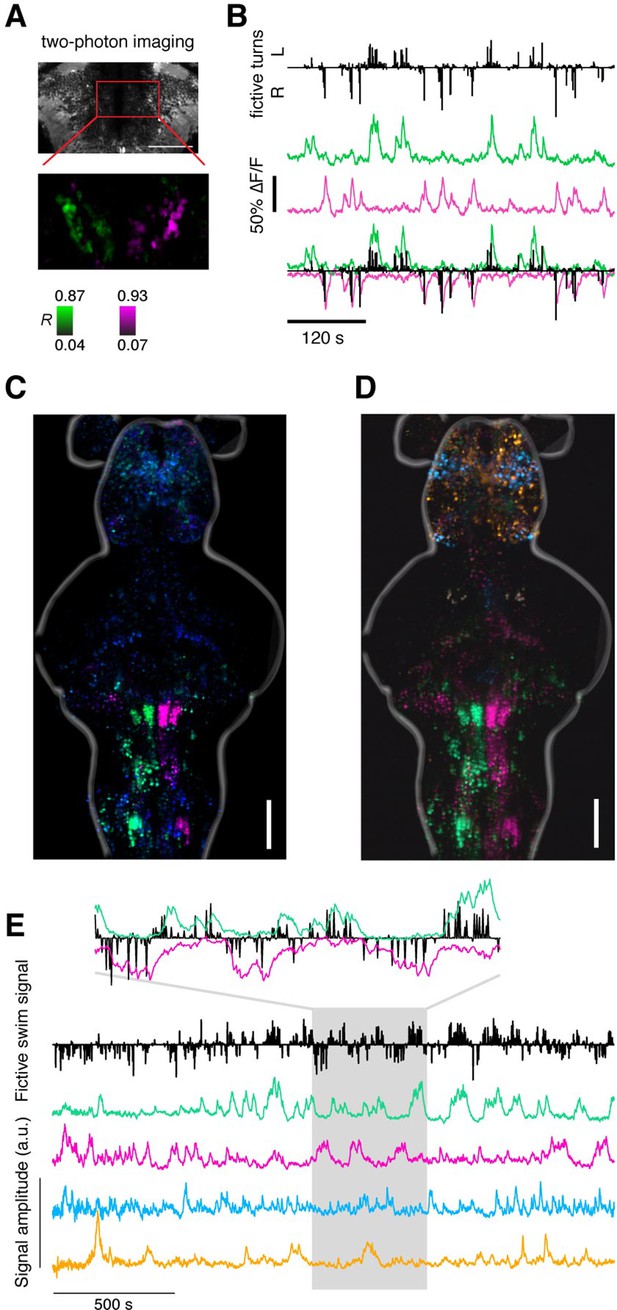
Recovering the ARTR using supervised and unsupervised methods.
(A) Two-photon imaging during fictive behavior recovers the ARTR in one plane by correlating every pixel to a seed voxel. This antiphasic correlation pattern was observed in all fish imaged, including fish used for neurite tracing and neurotransmitter analysis, N = 69 total. (B) Fictive turns (black bars) and neural activity (average over magenta and green populations) shows the correlation between two-photon ARTR signal and turning behavior. (C) Map derived from regression analysis relating behavioral parameters (turn direction and amplitude) to neuronal responses (see Figure 2 and 'Materials and methods'), conventions as in Figure 2. (D) Maximum projection maps derived from Independent Component Analysis (ICA), performed as described in Freeman et al. (2014). Analysis was applied to voxel-wise time series data using 100 principal components and 20 independent components. Spatial maps for 4 out of 20 components shown here, combined into a composite color image by scaling amplitude to a (black, red/green/cyan/magenta) color range, separately for each of the 4 components, and then computing a maximum over the vertical dimension. Location of the ARTR is consistent with that from Freeman et al. (2014) and Ahrens et al. (2013). One representative fish is shown, but the ICA analysis was able to recover the ARTR consistently in 5 fish tested. (E) Temporal components for the same ICA analysis shown in panel (D). Fictive swim signal as defined in Figure 2. The four colored traces correspond to the four spatial maps in panel (D). Inset highlights components (magenta and green) that recover a region including the ARTR; one signal inverted to emphasize correspondence with behavior.
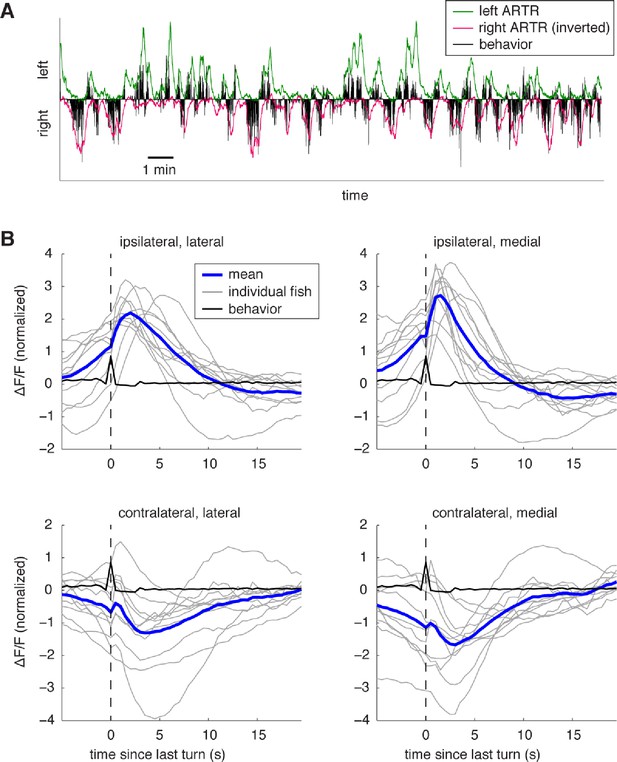
Dynamics of ARTR activity during behavior.
(A) Example of ARTR activity during a whole-brain recording with fictive behavior. During left (right) turns, the left (right) ARTR is activated. The cells in each of the two ARTR hemispheres were averaged to obtain the respective activity traces. (B) Turn-triggered averages of ARTR activity shows the neuronal activation during turns. Only turns that were followed by 5 s of no turning were included. Fluorescence traces were z-scored before averaging. Individual fish are shown in gray, average in blue, and behavior in black. The ARTR clusters ipsilateral to the turn direction are activated at the time of the turn, and calcium fluorescence peaks about 2 s after the turn, then decays on a timescale of 5–10 s. The lateral ARTR cluster appears to have slower dynamics than the medial ARTR cluster, although differences in calcium buffering may play a role in the difference in the fluorescence traces. Activity in the contralateral ARTR decreases after a turn.
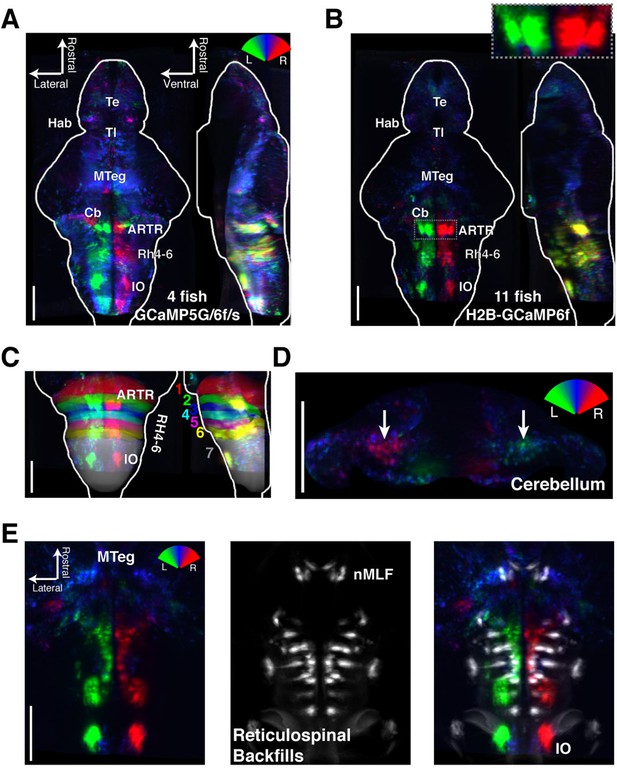
Functional anatomy of brain regions correlated with spontaneous behavior.
Activity patterns consistently observed across fish highlighted by registering multiple fish to the Z-Brain atlas and averaging functional signals (see Supplementary methods). (A) Average functional stack resulting from Tg(elavl3:GCaMP6f) (N=2), Tg(elavl3:GCaMP5G) (N=1) and Tg(elavl3:GCaMP6s) larvae (N=1). Color represents tuning to fictive turning as in Figure 2. (B) Average functional stack resulting from Tg(elavl3:H2B-GCaMP6f) larvae (n=11). (C–E) Anatomical analyses of the average Tg(elavl3:H2B-GCaMP6f) maps in (B). (C) The positioning of hindbrain within the rhombomeres. (D) Untuned and some more weakly direction selective signals observed in the cerebellum. (E) Virtual colocalization comparing the position of ventral hindbrain and midbrain tegmentum (M-Teg) signals with the reticulospinal system. R2 = 0…0.12 (A–C), 0…0.06 (D–E). Scale bars, 100 μm. (Te) telencephalon; (Hab) habenula; (Tl) torus longitudinalis; (Cb) cerebellum; (ARTR) anterior rhombencephalic turning region; (Rh4-6) rhombomeres 4-6; (IO) inferior olive; (nMLF) nucleus of the medial longitudinal fasciculus.
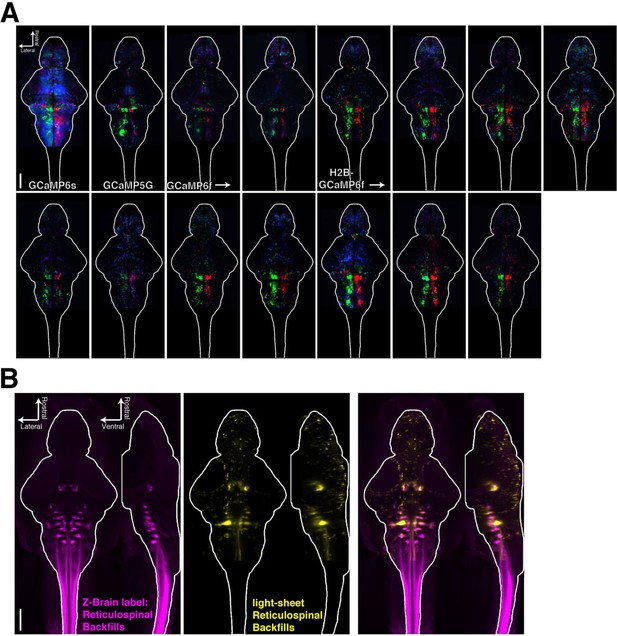
Registering brains to the Z-Brain atlas.
(A) Functional stacks from 15 fish aligned to the Z-Brain atlas. (B) Comparison of the reticulospinal backfill label in the Z-Brain atlas, with the mean reticulospinal backfills from 4 fish imaged by light sheet microscopy and aligned to the Z-Brain atlas using the Tg(elavl3:H2B-GCaMP6f) transgenic fish line.
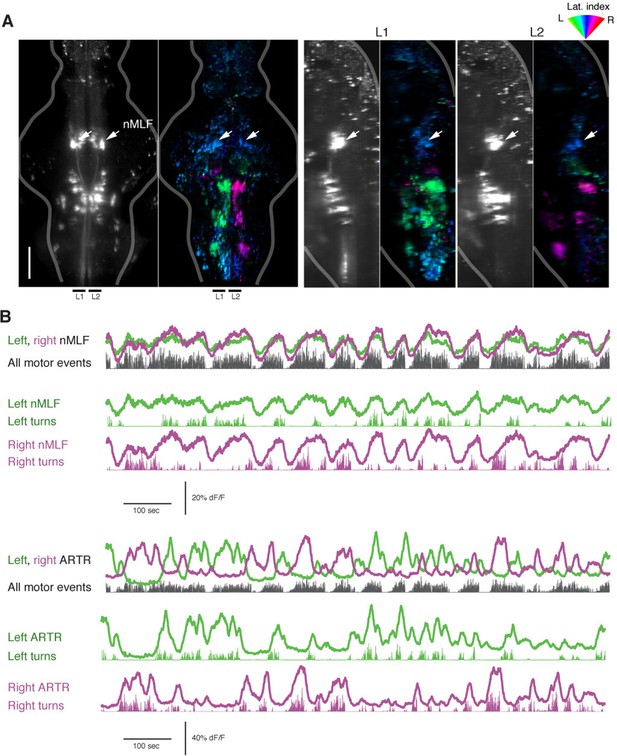
The nMLF is correlated with swim amplitude but not direction.
(A) Left, light-sheet micrograph of the backfilled reticulospinal system (grayscale, dextran-conjugated TxRed, see 'Materials and methods') and GCaMP6f regression map from the same fish. Right, sagittal views of the same fish, projected across the regions L1 and L2, as indicated to the left. Arrows in all maps point to the overlap between zones of blue-tuned regions in the functional brain maps and the backfilled nMLF neurons. The nMLF is activated by swimming, but not strongly tuned to direction. (B) Top, fluorescence time series for left and right nMLF ROIs with corresponding fictive behavior. Bottom, for comparison to nMLF tuning, fluorescence time series for left and right ARTR ROIs with corresponding fictive behavior.
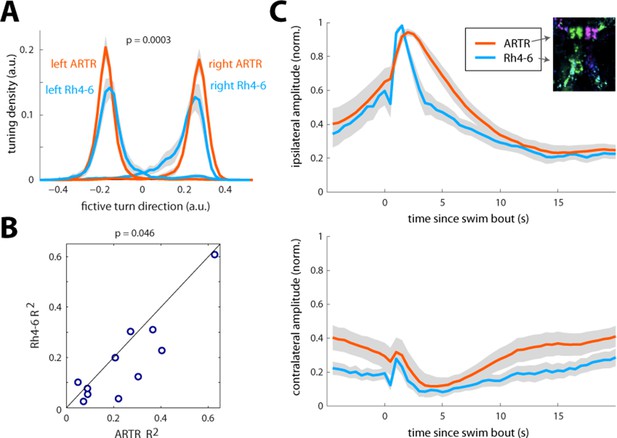
Comparison of ARTR and Rh4-6 dynamics, tuning, and predictiveness of future behavior.
(A) Tuning to turn index of ARTR cells vs. Rh4-6 cells. The histogram of laterality index across the ARTR and Rh4 populations shows that the ARTR is significantly shifted to larger indices. N = 11 fish. (B) The ARTR is more predictive of future turn direction than Rh4-6 cells. Since ARTR activity decays slowly, and the ARTR is able to bias turn direction (Figure 4), it is expected that future turn direction can be predicted from ARTR activity. To assess this, mean left and right ARTR activity, and mean left and right Rh4-6 activity, both 1 s prior to a turn, were regressed against turn direction. Only turns with 3 s of no prior motor events were included to prevent trivial correlations. Predictive power was assessed by R2 of the regression. The scatterplot of R2 from the ARTR prediction and Rh4 prediction shows that the ARTR is a stronger predictor of future turn direction. (C) Temporal dynamics of the ARTR and Rh4-6 cells. Average fluorescence traces for ARTR (red) and Rh4-6 (blue) neurons triggered on turns in the ipsilateral (top) or contralateral (bottom) direction. The ARTR decays on a longer timescale following an ipsilateral turn. In addition, both the contralateral ARTR and the contralateral side of Rh4 neurons decrease in activity following an ipsilateral turn, with a slight delay compared to ipsilateral neuron activation. Only turns with a subsequent period of at least 10 s with no motor events were included to be able to observe the decay dynamics. ARTR analyses use pooled data from both medial and lateral clusters (shown separately in Figure 2—figure supplement 3).
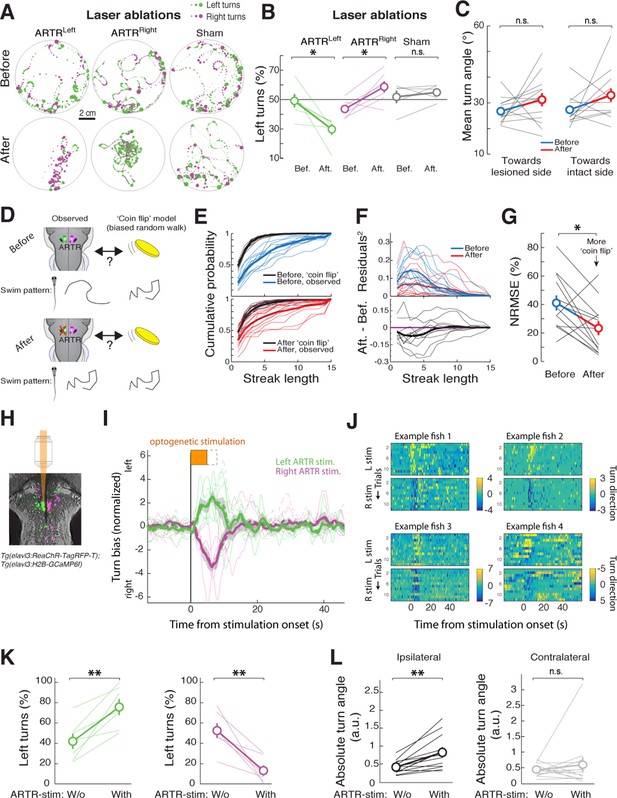
The ARTR biases turn direction.
(A,B) Unilateral laser ablation of a subset of cells in the ARTR reduces ipsilateral turns. (A) Example swim trajectories, shown over a subset of the duration of the experiment, and (B) summary of turning behavior before and after laser ablation of cells in the left medial cluster (green) or right medial cluster (magenta) of the ARTR. Gray, data from sham ablations of hindbrain neurons outside of the ARTR. Only events occurring more than 1 cm away from the wall were analyzed. (C) Mean turn angle to the lesioned or intact side, before and after ablation. Although the relative frequency of turns to the ablated side decreases (B), fish remain capable of executing turns of normal magnitude to the lesioned side, with no significant difference in mean turn angle between pre- and post-ablation conditions. (D) Schematic of hypothesized changes to swim structure, assuming that the ARTR is involved in setting correlational patterns. Before and after ablation, turn patterns will be compared to a ‘coin flip’ model that emits turns to the left and right randomly but with some bias equal to the observed data. (E) Empirical cumulative distribution functions (CDFs) of streak length before (blue, top) and after (red, bottom) ablation, compared to model fish executing turns at random without history dependence but with overall turn bias matched to each individual fish (black). Streak length post-ablation appears distributed more like 'coin flips'. (F) Top, quantification of the squared residuals between each individual fish CDF and its matched 'coin flip' CDF before (blue) and after (red) ablation. Bottom, the difference between each respective before and after curve reveals a shift toward the 'coin flip' distribution for the majority of fish. (G) Summary of the normalized root-mean-square error (NRMSE) quantifying goodness-of-fit between the observed streak distributions and their matched random model distributions. After ablation, turning becomes more ‘coin flip’-like and thus history dependence is reduced. (H–L) Optogenetic stimulation of the ARTR elicits ipsilateral turn biases. (H) The ARTR was functionally identified in double transgenic fish Tg(elavl3:H2B-GCaMP6f;elavl3:ReaChR-TagRFP-T) and a medial ARTR cluster was unilaterally stimulated. Gray, expression of ReaChR-TagRFP-T; green and magenta, functionally identified ARTR from this example fish based on correlational map. (I) Ipsilateral turn bias increases during optogenetic stimulation (solid lines, 5 s stimulation; dotted lines, 8 s stimulation, N = 7 fish, 'Materials and methods'). (J) Results from example fish show the reproducibility of stimulation effect across trials. Turn direction is normalized to time-averaged turn direction pre-stimulation. (K) Summary of the change in bias quantified for each fish, showing that optogenetic stimulation results in a bias toward ipsilateral turns. (L) Summary of the change in absolute fictive turn angle during stimulation, showing that ipsilateral turn angle increases and contralateral turn angle remains unchanged. n.s., no significance; (*) p<0.05; (**) p<0.01 (paired signed rank test). All error bars are mean ± SEM across fish.
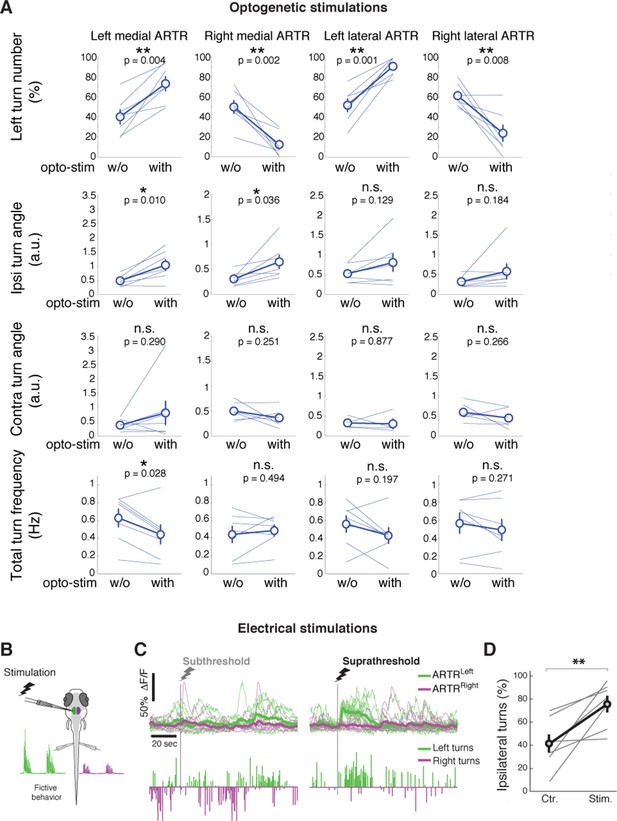
Detailed analysis of ARTR stimulations.
(A) Summary of changes to left turn bias, ipsilateral turn angle, contralateral turn angle, and turn frequency during optogenetic stimulation of the left medial, right medial, left lateral, and right lateral ARTR clusters. N =7 fish. Circle and error bars are mean ± SEM across fish. All p-values from a paired signed rank test. (**) p<0.001, (*) p<0.05. (B–D) Electrical stimulation of the ARTR enhances ipsilateral turns. (B) Schematic of the experimental setup for electrical stimulation. (C) Examples and (D) summary of turning behavior without and with suprathreshold electrical stimulation of the medial ARTR cluster on one side. N = 7 fish. Circles and error bars are mean ± SEM across fish. (**) p=0.011, rank sum test.
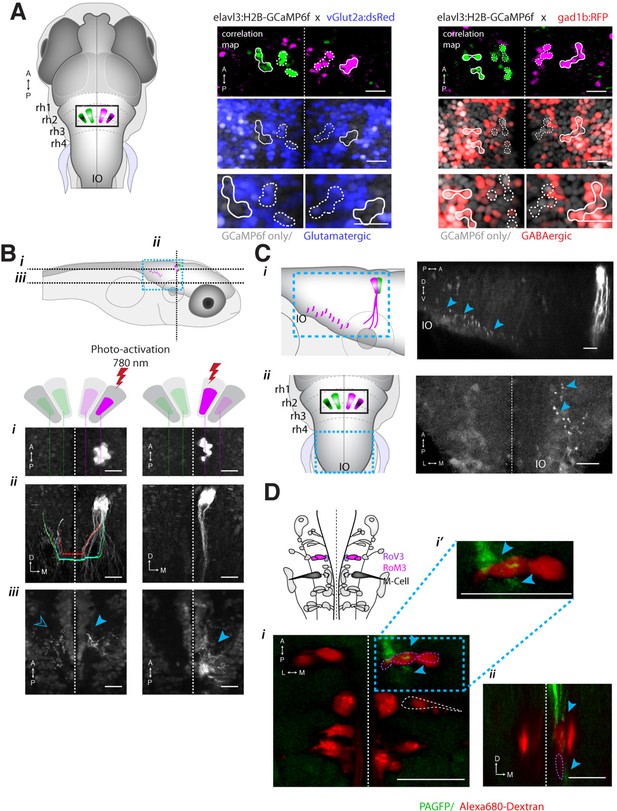
ARTR anatomy suggests mutual inhibition and connections to premotor neurons.
(A) The medial ARTR is glutamatergic and the lateral ARTR is GABA-ergic. Left, Anatomical diagram showing the approximate location of the ARTR (black box) in rhombomeres 2–3. Green and magenta represent clusters influencing left and right turns, respectively. Right, The ARTR was functionally identified in two-photon imaging sessions ('Materials and methods') in transgenic fish expressing H2B-GCaMP6f in most neurons and a red indicator either in the glutamatergic (left) or in the GABAergic (right) neurons. Overlaying the functional maps (top) in which the ARTR cells are identified by correlation reveals that the center clusters (dotted outlines) are glutamatergic and the lateral clusters (solid outlines) are GABAergic. For the vGlut experiments, N = 25 fish; GAD experiments, N = 11 fish; one representative fish shown for each. Scale bars, 20 μm. (IO) inferior olive; (rh1-4) rhombomeres 1-4. (B) The lateral ARTR projects contralaterally. Top, Anatomical diagram showing the approximate location of the planes shown below in (i), (ii), and (iii). The ARTR was identified as in (A) in Tg(H2B:GCaMP6f; α-tubulin-PAGFP) fish and PA-GFP was activated specifically in ARTR neurons of either the lateral cluster (left panels) or medial cluster (right panels). Projections were traced, revealing that the GABAergic cells of the lateral cluster cross the midline (dotted white line) toward the contralateral clusters (ii, left). The medial glutamatergic clusters project ventrally and ipsilaterally but were not found to cross the midline (ii and iii, right). Solid blue arrowheads, neurites in the hemisphere ipsilateral to the activated ARTR. Open blue arrowhead, neurites in the hemisphere contralateral to the activated ARTR. (C) Cells of the medial ARTR project to the ipsilateral IO. (i) The ARTR was functionally identified and photoactivated as in (B) in fish expressing PA-GFP and a red calcium indicator (Tg(elavl3:jRCaMP1a)). Left, schematic of the ARTR and PA-GFP-positive neurites projecting from the medial ARTR cluster and terminating in the IO (pink). Blue dashed rectangle represents the location of the region shown on the right. PA-GFP positive terminals are observed in the ipsilateral IO (blue arrowheads) (ii) Left, schematic showing the location (blue dashed rectangle) of (right) the top-down confocal image of the terminals shown in (i). (D) Cells of the medial ARTR send projections to a region nearby reticulospinal neurons. Top left, schematic of the reticulospinal system, adapted from Orger et al. (Orger et al., 2008), with RoV3, RoM3 and the Mauthner cell highlighted (Mauthner cell out of plane). (i) PA-GFP positive neurites (blue arrowheads) shown nearby RoV3 / RoM3 in fish where the reticulospinal neurons were retrogradely labeled with Alexa-680-dextran (red); panel (i’) shows a magnification of the boxed region. (ii) A coronal view of the two cells (RoV3/RoM3) shown in (i) and inset. Scale bars, 20 μm. A, anterior; P, posterior; L, lateral; M, medial; D, dorsal; V, ventral.
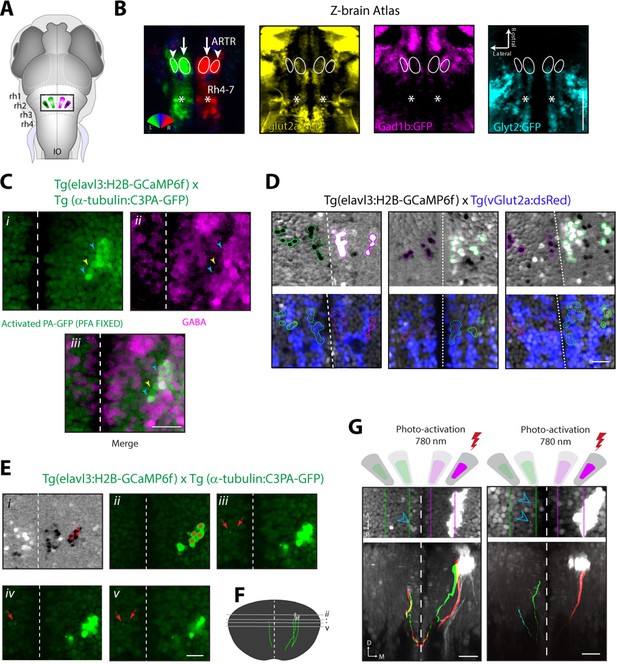
Cells of the lateral ARTR are inhibitory and project to the contralateral hindbrain.
(A) Schematic of a 6 dpf zebrafish brain indicating the approximate location of the ARTR in the dorsal hindbrain. (B) Virtual colocalization comparing the location of the ARTR and caudal hindbrain signals relative to Z-Brain neurotransmitter labels. (C) Cells of the lateral ARTR are still marked with photoactivated PA-GFP after fixation in 4% PFA-PBS. N = 12 fish, one representative fish shown. (Ci–Ciii) A majority of the GFP-positive cells are reactive with GABA primary antibody (cyan arrows), with a minority of exceptions (yellow arrows). (D) Three additional example fish showing vGlut2a-expressing neurons enriched in the medial ARTR clusters. Top row, correlation maps of ARTR activity. Bottom row, vGlut2a expression. (Ei) Correlation map of ARTR activity. Cells of the right lateral cluster were identified (red asterisks) and photoactivated (red asterisks). (Eii–Ev) Four different slices (dorsal to ventral) showing the photoactivated cells (right of the midline) and PA-GFP-positive processes on the contralateral side (red arrows). N = 37, one fish shown. (F) Reconstruction of traced photoactivated neurites (not all neurites were traced). The approximate levels of the slices in Eii–Ev are indicated. The cells project dorso-ventrally and turn toward the midline, at which point they could not be followed anymore in this example. Processes reappear on the contralateral side. In this example, the traced neurite terminates in the vicinity of the contralateral medial ARTR cluster. The vertical dimension of the ARTR is 83 ± 8 μm (mean ± SD, 6 fish). (G) Two additional example fish showing projections from the lateral ARTR cluster to the contralateral ARTR cluster. Note that only a subset of ARTR neurons are activated and that the entire ARTR structure spans approximately 80 microns along the dorso-ventral axis. Out of 10 fish manually traced, all 10 had detectable projections from the lateral cluster to the medial cluster on the contralateral side, and 5 had detectable projections to the lateral cluster on the contralateral side. Scale bars: 20 μm. Dashed white lines, brain midline.
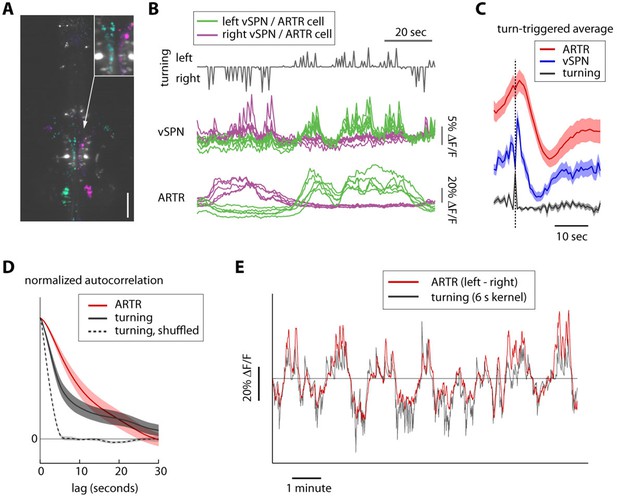
Timescales of reticulospinal, ARTR and turn state correlations.
(A) Ventromedial cells (vSPNs) in the hindbrain reticulospinal system (identified via spinal backfills, white) are correlated with turning behavior. Cyan and magenta show GCaMP6 voxels with laterality indices to the left and right, respectively. N = 4; one fish shown. Scale bar, 100 μm. (B) Example traces from 10 vSPNs (middle) and 10 ARTR neurons (bottom) during fictive swimming. ARTR neurons are active on longer timescales than vSPNs. (C) Quantification of ARTR (red) and vSPN (blue) response dynamics triggered on turns (gray). Only turns that were not followed by another turn in the same direction for at least 5 s were included to isolate ARTR dynamics from correlation in behavior. The ARTR signals are slower than the vSPN signals (as measured with the calcium indicator), and peak about 1.5–2 s after a turn, compared to a near-instantaneous peak in the vSPN signals, suggesting feedback from (peripheral) motor circuits to the ARTR. ARTR analyses use pooled data from both medial and lateral clusters (shown separately in Figure 2—figure supplement 3). (D) Normalized autocorrelation of ARTR signals (red), convolved turn laterality (gray), and convolved turn laterality after shuffling turn identity (dotted). ARTR activity and turning is correlated on similarly slow timescales. The length of the shuffled correlation reflects the width of the convolution kernel. (E) Convolved turning behavior (gray) matches the instantaneous difference between left and right ARTR fluorescence. ARTR analyses use pooled data from both medial and lateral clusters.
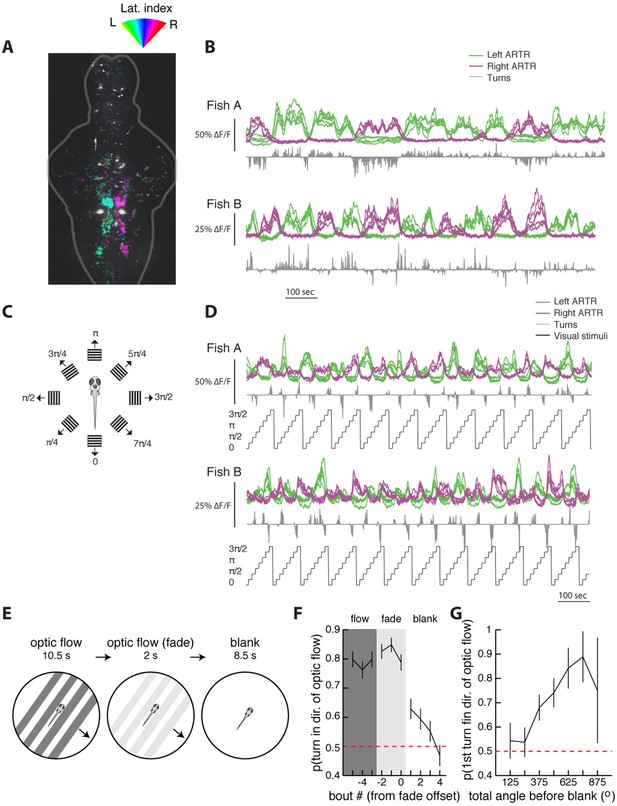
The ARTR is recruited by whole-field motion.
(A) Laterality tuning map for one fish, showing the characteristic long-timescale correlations with fictive behavior (B). This map is used to choose left and right ARTR ROIs that are used to assay ARTR responses to different directions of whole-field motion (C). (D) Responses of the ARTR ROIs to the eight directions of motion indicated in (C). This periodic motion paradigm drives turning strongly for orthogonal directions of motion and drives ARTR activity periodically, with the left ARTR activated for stimuli oriented to the left and the right ARTR activated for stimuli oriented to the right. (E) Experimental paradigm designed to test the effect of ARTR activation by whole-field motion. Freely swimming larvae are presented with drifting gratings moving orthogonal to the body axis, either to the left or the right, depending on the trial. These gratings are presented with closed-loop feedback such that they are always locked to the fish’s body axis even as the fish swims around the petri dish. This stimulus drives turning in the direction of motion. After 10.5 s of optic flow, the gratings fade away linearly over 2 s, and behavior is monitored under subsequent spontaneous conditions for 8.5 s. (F) The probability of turning in the direction of flow reaches a maximum during the flow and fade epochs but stays elevated above chance for three bouts into the null epoch. (G) The direction of the first swim bout following visual stimulation is strongly related to the amount of turning during optic flow. The more the fish turns during optical stimulation, the higher the chance that the first swim bout in the blank period is in the same direction (22 fish) Figure 6G–H.
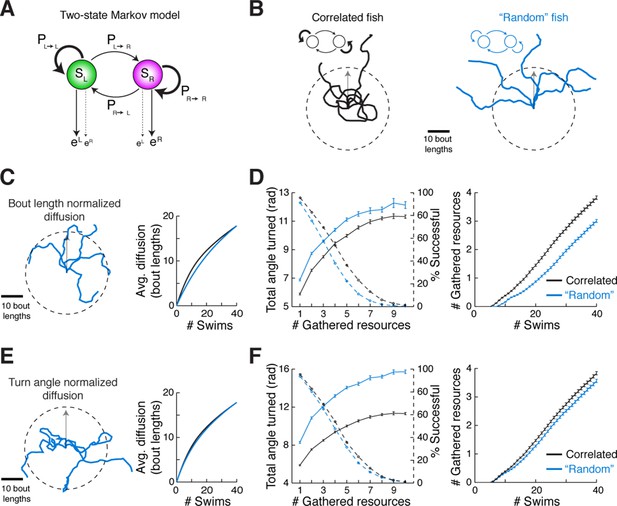
Correlated turn states may underlie efficient local exploration (A) Spontaneous turn states are well-characterized by a two-state Markov model (Figure 6—figure supplement 1).
In an average model fit, fish in the left state, SL, are much more likely (~90%) to turn left (eL) than right (eR), and vice-versa. And fish in SL or SR tend to return to SL or SR, respectively, after a turn. (B) Left, black, Five swim trajectories generated with a Markov model matching the statistics of acquired swim data (see fish 16, Figure 6—figure supplement 1, Ptransition = [PL►L PL►R PR►L PR►R] = [0.86 0.14 0.15 0.85]). Right, blue, five swim trajectories generated with a Markov model randomly emitting left and right turns (all Ptransition = 0.5). Notice that the unadjusted 'random' fish diffuses farther from the given starting position. The dotted circle represents the mean diffusion distance for the correlated model fish. All trajectories begin at the center of the circle and facing in the direction of the arrow. (C) Left, five example trajectories from the 'random' model fish after average diffusion has (right) been matched to the correlated fish by decreasing bout distance. (D) Plots of exploration efficiency for the 'random' model fish normalized by bout distance. Left, in this local regime, the 'random' model fish must turn more (16.9% more for 1 resource, p < 10–9; p = 0.004 for 10 resources, two-tailed t-test) and (right) execute more swim bouts (21.3% fewer resources after 40 swims, p=<10–9, two-tailed t-test) than the correlated model to collect randomly distributed virtual resources. Left, dashed lines, plots showing the proportion of simulated trajectories able to gather the indicated number of resources after 40 swims. Error bars are SEM, see Figure 6—figure supplement 1E,F; 'Materials and methods'. (E) Left, five example trajectories from the 'random' model fish after average diffusion has (right) been matched to the correlated model fish by broadening the underlying turn angle distribution. (F) Plots of exploration efficiency for the 'random' model fish normalized by turn angle. Left, this 'random' fish must turn much more (40.8% more for 1 resource, p<10–9; p<10–9 for 10 resources, two-tailed t-test) and (right) execute more swim bouts (7.0% fewer resources after 40 swims, p=9.9 × 10–4, two-tailed t-test) than the correlated model to collect randomly distributed virtual resources. Left, dashed lines, plots showing the proportion of simulated trajectories able to gather the indicated number of resources after 40 swims. Error bars are SEM, see Figure 6—figure supplement 1G,H; 'Materials and methods'.
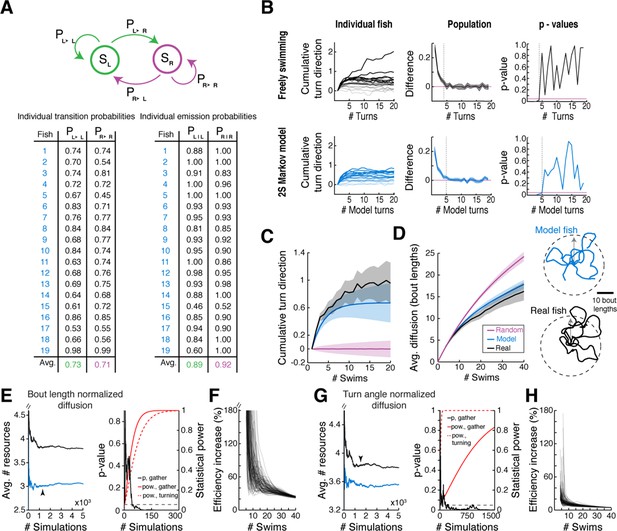
Validation of two-state Markov model.
(A) Table of the trained Markov model transmission and emission matrices for each fish in the data set. N = 19 fish. (B) Signed cumulative turn angle triggered on left->right or rig ht->left switch events for individual fish (left column) and change in signed turn angle across fish (middle column) for freely swimming fish (top row) and the output of the Markov model (bottom row). A p-value threshold of 0.05, for a signed rank test between the change in cumulative turn angle and a randomly turning fish (pink line, 0) reveals that average state length for the Markov model is the same as for freely swimming fish. (C) Direct comparison of streak statistics for a freely swimming fish and its corresponding Markov model output (fish 16 in (A); each fish generates a unique model output based on best-fit emission and transition probabilities). (D) Left, diffusion rate for the freely swimming and model data (as in C, left). Right, example trajectories from the same fish and model as to the left. Note the similarities in diffusion distance and curvature. (E) Left, plots of mean resources gathered after 40 swims over number of simulated trajectories for the correlated (blue) and bout length normalized 'random' model (black). Black arrowhead indicates the critical number of simulated trajectories (see 'Materials and methods'). Right, plots of two-tailed t-test p-value (black) for the number of resources gathered after 40 swims (see Figure 6D, right) and the corresponding statistical power (red) over number of simulated trajectories. Dashed red, statistical power over number of simulated trajectories for the total turn angle required for 1 resource. (F) Plots of % increase in resources gathered for the correlated model over the bout length normalized 'random' model at population scale (N = 106 simulated trajectories) for 100 different random resource distributions. (G) Left, plots of mean resources gathered after 40 swims over number of simulated trajectories for the correlated (blue) and turn angle normalized 'random' model (black). Black arrowhead indicates the critical number of simulated trajectories (see 'Materials and methods'). Right, plots of two-tailed t-test p-value (black) for the number of resources gathered after 40 swims (see Figure 6D, right) and the corresponding statistical power (red) over number of simulated trajectories. Dashed red, statistical power over number of simulated trajectories for the total turn angle required for 1 resource. (H) Plots of% increase in resources gathered for the correlated model over the turn angle normalized 'random' model at population scale (N = 106 simulated trajectories) for 100 different random resource distributions.
Videos
Spontaneous freely swimming behavior video of a freely swimming larval zebrafish, with turn direction coded by color (green = left; magenta = right) and turn amplitude coded by the size of the dot.
Fish was masked from background and smoothed to produce the representation on the top right.
Whole-brain imaging during spontaneous fictive behavior recorded in the light-sheet virtual reality setup.
Left: top projection of whole-brain ΔF/F. Top left: behavior represented by left and right turn amplitude. Right: projections of whole-brain ΔF/F, where the brain has been masked by the R2 volume, so that each voxel represents ΔF/F x R2, emphasizing neural activity in the regions identified by the regression analysis.
Analysis of imaging data Computational brain maps of the voxel-wise tuning to the laterality of turns.
Green signifies tuning to left turns; magenta to right turns; brightness codes for R2 of the model fit. Same data as Figure 2 but represented in three dimensions.
3D representation of Z-Brain atlas.
https://doi.org/10.7554/eLife.12741.016Additional files
-
Supplementary file 1
Example whole-brain data and analysis code (the data is on Amazon S3; the example analysis code links to the data).
- https://doi.org/10.7554/eLife.12741.029
-
Supplementary file 2
Table of brain areas with functional signals for the Z-Brain atlas.
- https://doi.org/10.7554/eLife.12741.030





