Tethering of an E3 ligase by PCM1 regulates the abundance of centrosomal KIAA0586/Talpid3 and promotes ciliogenesis
Figures
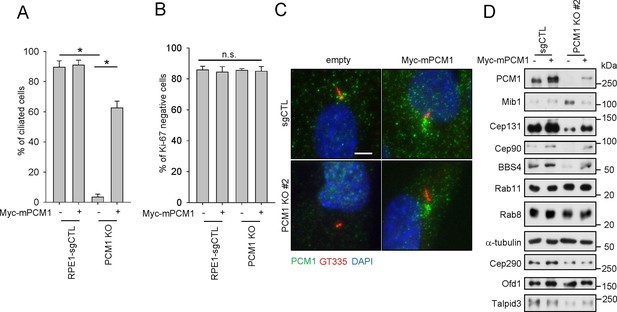
PCM1 regulates the abundance of centriolar satellite proteins and is essential for ciliogenesis. CRISPR/Cas9 deletion of PCM1 inhibits ciliogenesis.
(A) and (B) Control and PCM1 KO RPE1 cells were infected with empty or Myc-mPCM1 lentivirus for 72 hr and then serum starved for 48 hr. The cells were immuno-stained with GT335 and Ki-67 antibodies. (A) Ciliated cells and (B) Ki-67-negative cells were counted; n ≥ 100 per sample in three independent experiments. Error bars, SEM. *p<0.05 (C) Ectopic PCM1 can rescue the ciliogenesis defect of PCM1 KO RPE cells. Control and PCM1 KO RPE1 cells were infected with empty or Myc-mPCM1 lentivirus for 72 hr, serum starved for 48 h, and immuno-stained with antibodies against glutamylated tubulin (GT335, red), and PCM1 (green) and with DAPI (blue). Representative images are shown. Scale bar, 5 µm. (D) Altered levels of centriolar satellite proteins in PCM1 KO RPE1 cells. Control and PCM1 KO RPE1 cells were infected with control or Myc-mPCM1 lentivirus for 72 hr and subjected to western blot analysis with the indicated antibodies.
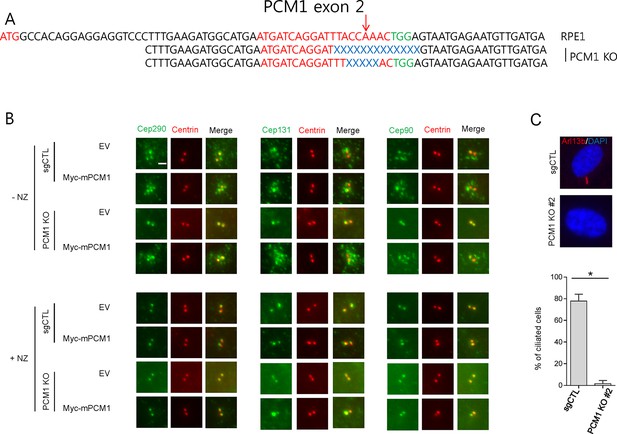
PCM1 gene-editing and the localization of Cep290, Cep131 and Cep90 in PCM1 KO cells.
(A) PCM1 genomic regions targeted by sgRNAs (sgPCM1) to generate PCM1 knock-out (KO) RPE1 cells were analyzed. The sequences were compared with wild type PCM1 genomic sequence. Red, blue and green indicates the target sequence, deletion mutation and PAM motif, respectively. Red arrow is the expected cleavage site. (B) Control and PCM1 KO RPE1 cells were infected by empty or Myc-mPCM1 lentivirus for 3days and and incubated with nocodazole, NZ (1 μg/ml) for 8 hr. Cell were then immunostained with Cep290/Cep131/Cep90 (green) and Myc (red). Scale bar, 2 µm. (C) Ciliation defect in PCM1 KO cells. The cells were immunostained with Arl13b antibody. Ciliated cells were counted; n ≥ 100 per sample in three independent experiments. Error bars, SD. *p<0.05
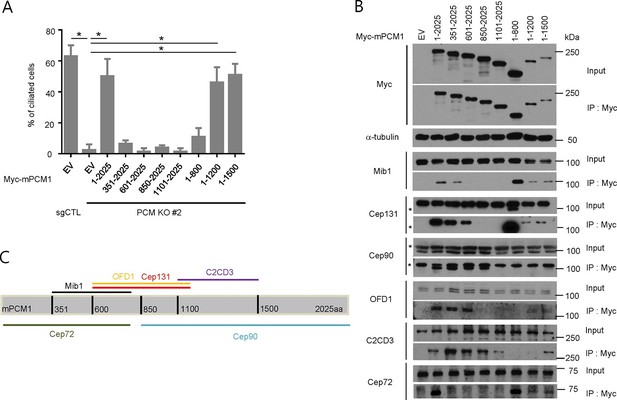
PCM1 promotes centriolar satellite organization and ciliogenesis through an amino-terminal domain.
(A) Mapping of PCM1 domains required to rescue the ciliogenesis defect in PCM1 null cells. Control and PCM1 KO RPE1 cells were infected with virus containing the empty vector (EV) or the indicated Myc-mPCM1 fragments. Cells were infected with lentivirus for 3 days and serum starved for 48 hr. The cells were immuno-stained with antibodies against IFT88, detyrosinated tubulin, and the Myc epitope. Ciliated cells were counted within the Myc-positive population. n ≥100 were counted per sample in two independent experiments. Error bars, SD. *p<0.05. (B, C) Identifying PCM1 domains required to interact with centriolar satellite proteins. (B) HEK293T cells were transfected with plasmids corresponding to the empty vector (EV) or the indicated Myc-mPCM1 fragments for 48 h, and lysates subjected to immunoprecipitation with anti-Myc antibody. The inputs and the immunoprecipitates were analyzed by western blotting with the indicated antibodies. Asterisks indicate non-specific cross-reactive bands. We surmise that the band detected by anti-Cep131 antibodies in cell extracts expressing fragment 1–800 is cross-reactive, as this species consistently migrates at a position distinct from Cep131. (C) Schematic representation of protein-protein interacting domains in PCM1 based on immunoprecipitation data in (B).
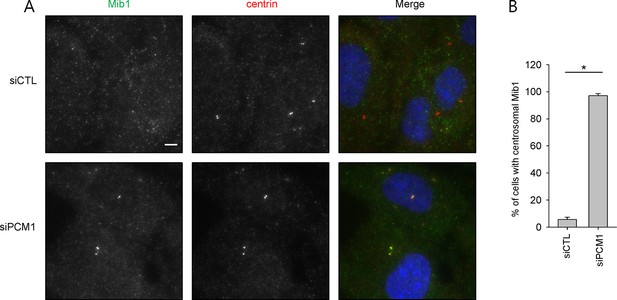
Centrosomal localization of Mib1 in PCM1 knock-down cells.
(A) and (B) RPE1 cells were transfected with control (siCTL) or PCM1 (siPCM1) siRNAs for 48 hr and immunostained with Mib1 (green), centrin (red) and DAPI (blue). (A) Representative images are shown. Scale bar, 20 µm. (B) Cells with centrosomal Mib1 were counted. N ≥ 100 per sample in two independent experiments. Error bars, SD. *p<0.05.
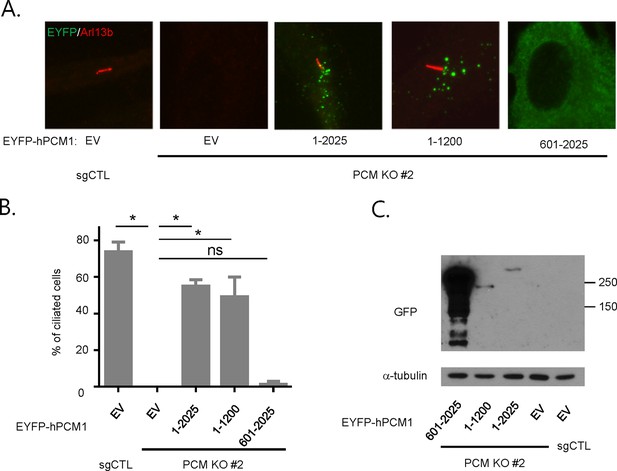
PCM1 promotes ciliogenesis through an amino-terminal domain.
(A, B) Mapping of human PCM1 domains required to rescue the ciliogenesis defect in PCM1 null cells. Control and PCM1 KO RPE1 cells were infected with virus containing the empty vector (EV) or the indicated EYFP-hPCM1 fragments. Cells were infected with lentivirus for 3 days and serum starved for 48 hr. The cells were immunostained with antibodies against Arl13b and EGFP. Ciliated cells were counted within the EYFP-positive population. n ≥100 were counted per sample in two independent experiments. Error bars, SD. *p<0.05. (C) Validation of expression of EYFP-hPCM1 by western blot analysis with the indicated antibodies.
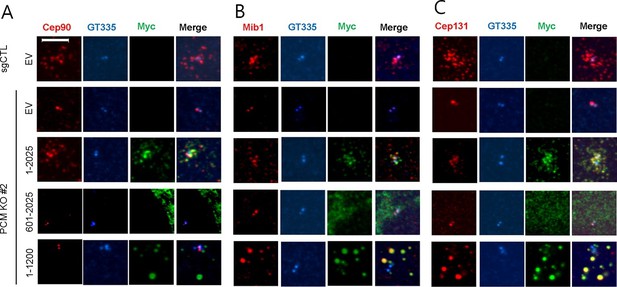
The amino-terminal domain of PCM1 recruits specific centriolar satellite proteins.
Control and PCM1 KO RPE1 cells were infected with lentivirus containing empty vector or the indicated Myc-mPCM1 fragments for 3days and stained with the antibodies against Myc, GT335, Cep90, Mib1, and Cep131. Diffuse cytoplasmic staining of PCM1 was observed with the fragment spanning residues 601–2025. Scale bar, 5 µm.
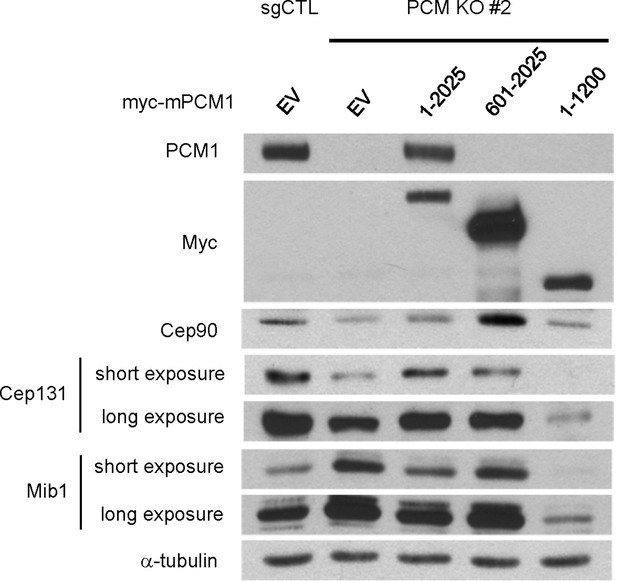
The amino-terminal domain of PCM1 recruits specific centriolar satellite proteins.
Control and PCM1 KO RPE1 cells were infected with lentivirus containing empty vector or the indicated Myc-mPCM1 fragments for 3 days and subjected to western blotting with the indicated antibodies.
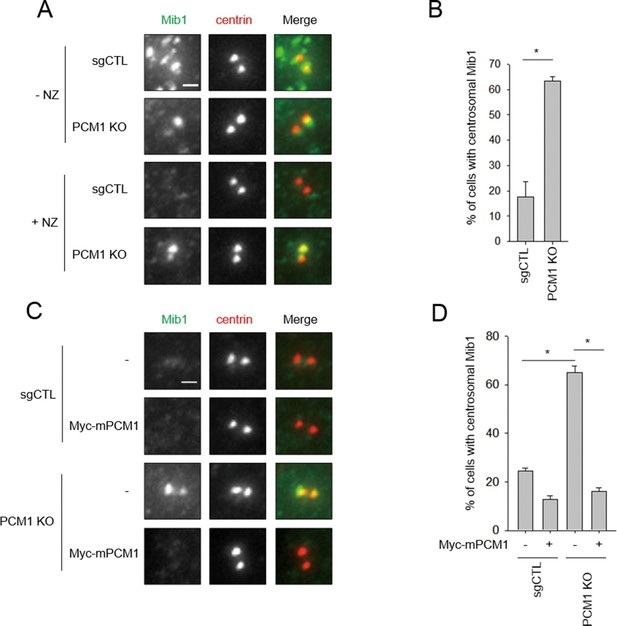
Mib1 re-locates to centrosomes in PCM1 null cells.
(A) Control and PCM1 KO RPE1 cell were incubated with DMSO or nocodazole (NZ; 1μg/ml) for 8 hr and then immuno-stained with Mib1 (green) and centrin (red) antibodies (A) and (B). Representative images of centrosomes are shown (A). Scale bar, 1 µm. (B) Quantitation of nocodazole-treated cells in panel A. Cells with centrosomal Mib1 signal were counted in the NZ-treated group. n ≥ 100 per sample in two independent experiments. Error bars, SD. *p<0.05. (C) Ectopic PCM1 expression in PCM1 KO RPE cells prevents centrosomal localization of Mib1. Control and PCM1 KO RPE1 cells were infected with empty or Myc-mPCM1 lentiviruses for 72 hr and incubated with NZ (1 μg/ml) for 8 hr (C) and (D). (C) Cells with centrosomal Mib1 signal were analyzed by immuno-staining with Mib1 (green) and centrin (red) antibodies. Representative images of centrosome are shown. Scale bar, 1 µm. (D) Quantitation of data in panel C. N ≥ 100 per sample in two independent experiments. Error bars, SD. *p<0.05.
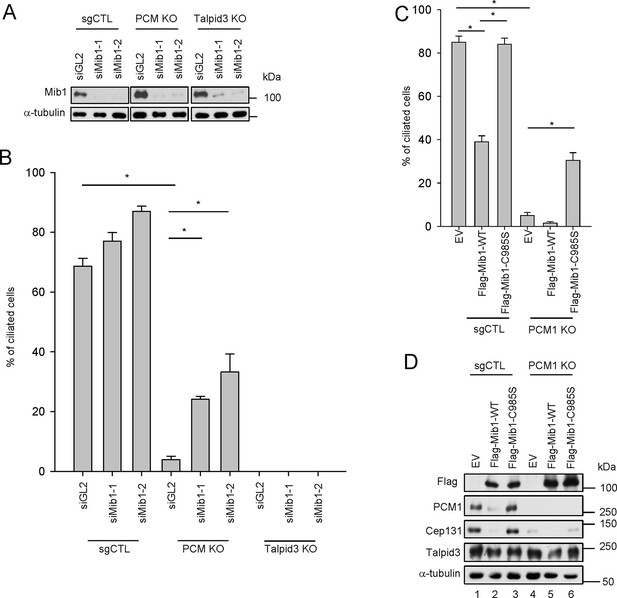
Mib1 is downstream of PCM1.
(A) Validation of siRNA knockdown of Mib1. Control (sgCTL), PCM1, and TALPID3 KO RPE1 cells were transfected with the indicated siRNAs corresponding to non-specific control (siGL2) or Mib1 (siMIB1-1 or siMIB1-2), and lysates were subjected to western blot analysis with the indicated antibodies. (B) Mib1 depletion partially rescues the ciliogenesis defect in PCM1 KO cells. Cell lines in panel A were transfected with siRNAs corresponding to non-specific control (siGL2) or Mib1 (siMIB1-1 or siMIB1-2) for 48 hr and serum starved for 48 hr. Ciliated cells (n ≥ 100 per sample in three independent experiments) were analyzed by immuno-staining with GT335. Error bars, SEM. *p<0.05. (C) E3 ligase activity of Mib1 underlies ciliogenesis defect in PCM1 KO cells. Control and PCM1 KO RPE1 cells were infected with empty (EV), Flag-Mib1-WT or Flag-Mib1-C985S lentiviruses for 72 hr and serum starved for 48 hr. Ciliated cells were analyzed by immuno-staining with GT335; n≥100 per sample were analyzed in two independent experiments. Error bars, SD. *p<0.05. (D) Control, PCM1, and Talpid3 KO RPE1 cells were transfected with siGL2, siMIB1-2, or siMIB1-3 for 48 hr and subjected to western blot analysis with the indicated antibodies.
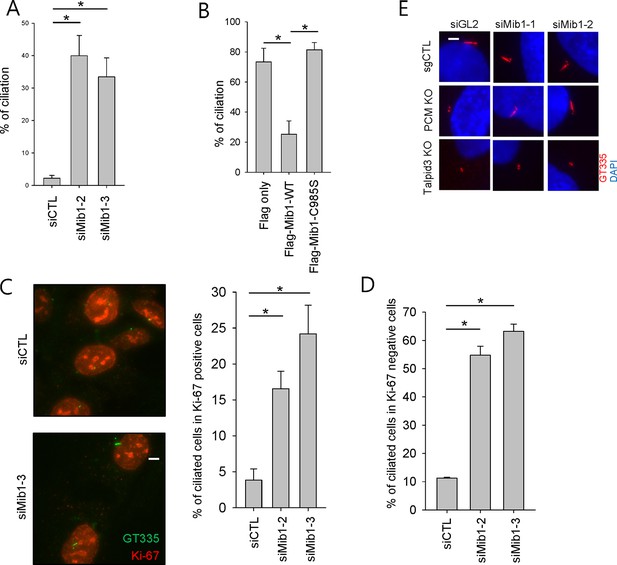
Mib1 suppress ciliogenesis.
(A) RPE1 cells were transfected with siCTL, siMIB1-2 or siMIB1-3 for 2 days. The ciliated cells were determined by immunostaining with GT335 antibody. N ≥ 100 per sample in three independent experiments. Error bars, SEM. *p<0.05. (B) RPE1 cells were infected with empty, Flag-Mib1-WT, or Flag-Mib1-C985S lentiviruses for 72 hr and serum starved for 48 hr. GT335-positive ciliated cells were counted. N ≥ 100 per sample in two independent experiments. Error bars, SD. *p<0.05. (C) and (D) RPE1 cells were transfected with siCTL, siMIB1-2 or siMIB1-3 for 2 days and immunostained with GT335 (green) and Ki-67 (red) antibodies. The ciliated cells were counted in Ki-67 (C) positive cells or (D) negative cells. N ≥ 100 per sample in two independent experiments. Error bars, SD. *p<0.05. (E) Mib1 depletion partially rescues the ciliogenesis defect in PCM1 KO cells. Control (sgCTL), PCM1, and TALPID3 KO RPE1 cells were transfected with the indicated siRNAs corresponding to non-specific control (siGL2) or Mib1 (siMIB1-1 or siMIB1-2) for 48 hr and serum starved for 48 hr. Ciliated cells were analyzed by immuno-staining with GT335.
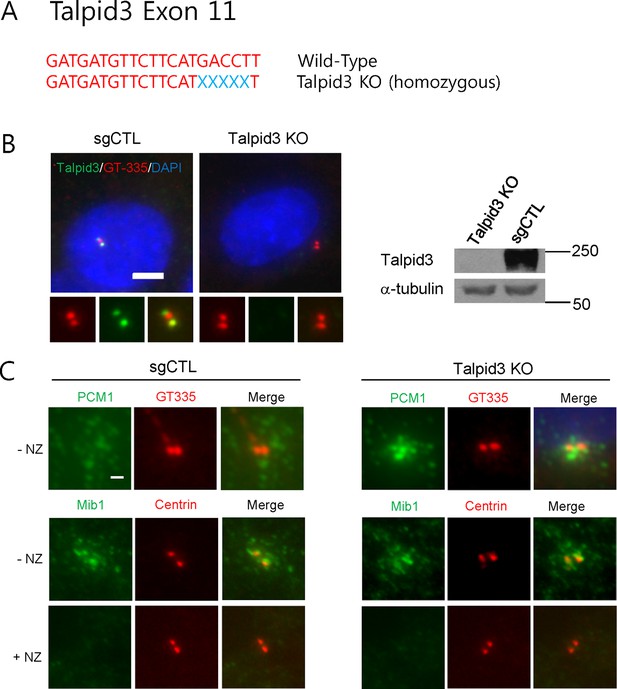
Generation of TALPID3 knock-out cell lines.
(A) Gene-editing with CRISPR/Cas9 was used to create the indicated mutations in RPE1 cells, the sequence for which is shown. (B) Talpid3 protein was undetectable by immunofluorescent detection and western blotting. Higher magnification views of centrioles are shown below the main panel in (B). Scale bar, 5 µm. (C) localization of PCM1 and Mib in TALPID3 KO cells. Control and TALPID3 KO RPE1 cells were incubated with or without NZ (1 μg/ml) for 8 hr. Cell were then immuno-stained with PCM1 (green), Mib1 (green), Centrin (red) and GT-335 (red). Scale bar, 2 µm.
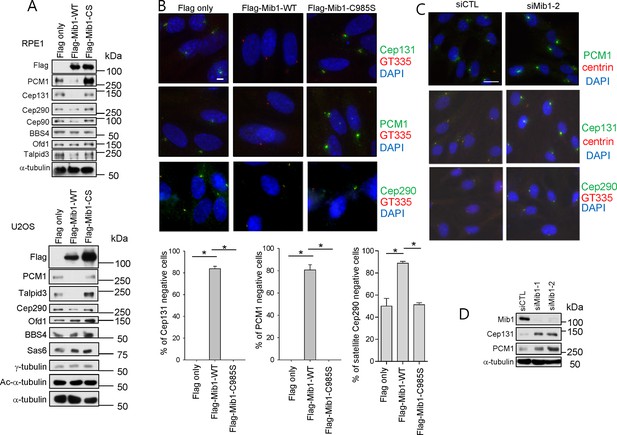
Mib1 regulates the stability of multiple centriolar satellite proteins.
(A) and (B) RPE1 or U2OS cells were infected with empty, Flag-Mib1-WT, or Flag-Mib1-C985S lentivirus for 3 days and subjected to (A) western blotting with the indicated antibodies or (B) immunostaining with DAPI (blue), GT335 (red), and Cep131 (green), CEP290 (green) or PCM1 (green). Scale bar, 5 µm. N ≥ 100 per sample in two independent experiments. Error bars, SD. *p<0.05. (C) and (D) RPE1 cells were transfected with siRNA for Mib1 and (C) immunostained with DAPI (blue), centrin (red), and Cep131 (green), CEP290 (green) or PCM1 (green) or subjected to (D) western blotting with the indicated antibodies.
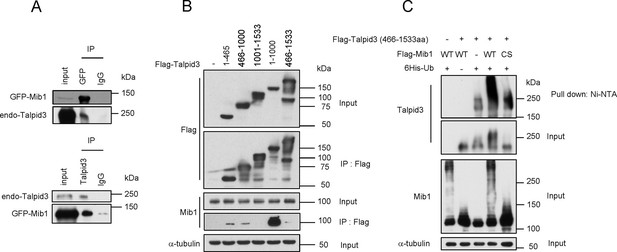
Mib1 promotes poly-ubiquitylation of Talpid 3.
(A) Mib1 interacts with Talpid3. GFP-Mib1 was expressed in HEK293T cells, lysates were subjected to immunoprecipitation with control IgG, anti-GFP, or Talpid3 antibodies, and western blot analysis was performed with the indicated antibodies. (B) Mapping of Talpid3 domains that interact with Mib1. HEK293T cells were transfected with control (-) or expression vectors encoding indicated Flag-tagged Talpid3 fragments for 48 h, and lysates were immunoprecipitated with anti-Flag antibodies. The input and immunoprecipitates were analyzed by western blotting with the indicated antibodies. (C) Ubiquitylation of Talpid3 depends on the integrity of the Mib1 Ring domain. 293T cells were transfected with plasmids expressing His-ubquitin, Flag-Mib1-WT, Flag-Mib1-C985S and/or Flag-Talpid3 (466-1533aa) for 2 days, and then purified using Ni-NTA resin to detect in vivo ubiquitylation. The inputs and the Ni-NTA Resin pull-down samples were analyzed by western blotting with the indicated antibodies.
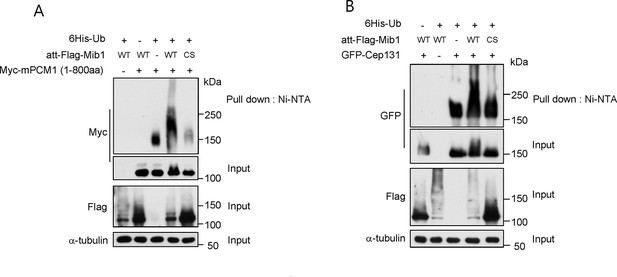
Mib1 ubiquitylates Cep131 and PCM1.
(A) and (B) HEK293T cells were transfected with vectors expressing His-tagged ubquitin, Flag-Mib1-WT, Flag-Mib1-C985S and/or, Myc-mPCM1 (residues 1–800) or Flag-Cep131 for 2 days, and then subjected to in vivo ubiquitination assays in which conjugates were purified using Ni-NTA resin. Input and the Ni-NTA purified samples were analyzed by western blotting with the indicated antibodies.
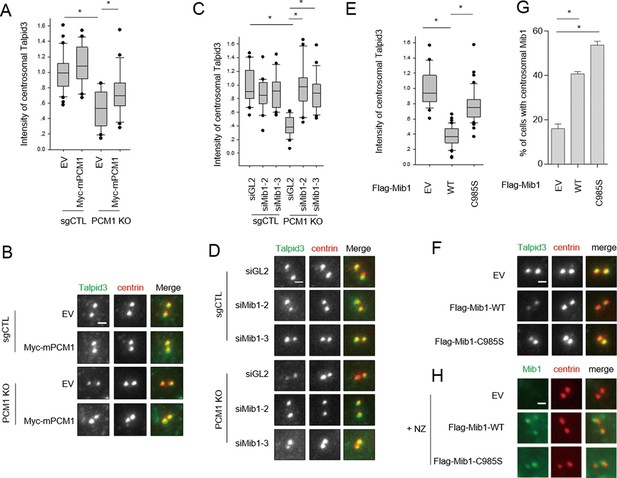
PCM1 regulates Talpid3 abundance by sequestering Mib1.
(A) and (B) Ectopic PCM1 expression in PCM1 KO cells stabilizes Talpid3 at centrioles. Control and PCM1 KO cells were infected with empty or Myc-mPCM1 lentivirus for 3 days. (A) Centrosomal Talpid3 intensity was measured in G1 phase cells by immuno-staining with anti-Talpid3 (green) and centrin (red) antibodies. N ≥ 20 per sample in two independent experiments. Error bars, SD. *p<0.05. (B) Representative images are shown. Scale bar, 1 µm. (C) and (D) Depletion of Mib1 in PCM1 null cells stabilizes Talpid3 at centrioles. Control and PCM1 KO RPE1 cells were transfected with siGL2, siMIB1-2 or siMIB1-3 for 2 days. (C) Centrosomal Talpid3 intensity in G1 phase cells was measured by immuno-staining with anti-Talpid3 (green) and centrin (red) antibodies. N ≥ 20 per sample in two independent experiments. Error bars, SD. *p<0.05. (D) The representative images are shown. Scale bar, 1 µm. (E) and (F) Destabilization of Talpid3 at the centrosome by Mib1 is dependent on a functional RING domain. RPE1 cells were infected with empty, Flag-Mib1-WT or Flag-Mib1-C985S lentiviruses for 72 hr. (E) Centrosomal Talpid3 intensity in G1 phase cells was measured by immunostaining with Talpid3 (green) and centrin (red) antibodies. N ≥ 20 per sample in two independent experiments. Error bars, SD. *p<0.05. (F) The representative images are shown. Scale bar, 1 µm. (G) and (H) Centrosomal localization of Mib1 in cells ectopically expressing Mib1. (G) Cells were analyzed by immuno-staining with Mib1 (green) and centrin (red) antibodies. Cells with centrosomal Mib1 were counted. N ≥ 100 per sample in two independent experiments. Error bars, SD. *p<0.05. (H) Representative images are shown. Scale bar, 1 µm.
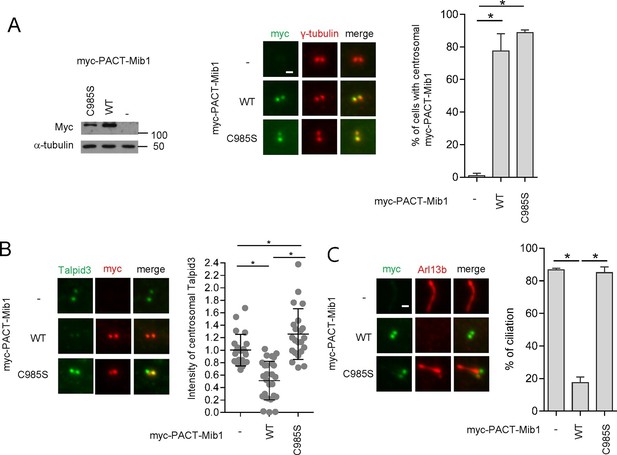
Centrosomal targeting of Mib1 destabilizes Talpid3 and inhibits ciliogenesis.
(A) (Left) RPE1 cells were infected with empty (-), myc-PACT-Mib1-WT, or myc-PACT-Mib1-C985S lentiviruses for 72 hr and western blotting performed to detect the fusion protein. (Middle) Visualization of centrosomal localization of Mib1 in cells infected as in (A). Myc-PACT-Mib1 was detected by immuno-staining with anti-myc (green) and γ-tubulin (red) antibodies. (Right) Cells with centrosomal Mib1 were counted. N ≥ 100 per sample in two independent experiments. Error bars, SD. *p<0.05. (B) Destabilization of Talpid3 at the centrosome by centrosomal targeting of Mib1. Centrosomal Talpid3 intensity in G1 phase cells was measured by immuno-staining cells exclusively exhibiting centrosomal myc-PACT-Mib1 with anti-Talpid3 (green) and myc (red) antibodies. N ≥ 40 per sample in two independent experiments. Error bars, SD. *p<0.05. (C) Ciliated cells exhibiting centrosomal myc-PACT-Mib1 (as in panel B) were analyzed by immuno-staining with myc (green) and Arl13b (red) antibodies; n≥100 per sample were analyzed in two independent experiments. Error bars, SD. *p<0.05.
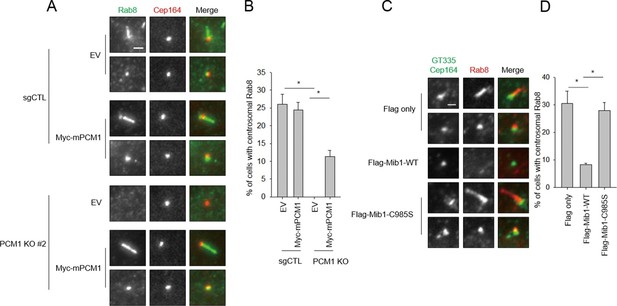
Mib1 expression or ablation of PCM1 blocks ciliogenesis at an early stage prior to Rab8 recruitment.
(A) PCM1 null cells are defective in recruitment of Rab8 to the mother centriole. (B) Quantitation of Rab8 in PCM1 null cells. (A) and (B) Control and PCM1 KO RPE1 cells were infected with vector control (EV) or Myc-mPCM1 lentiviruses for 72 hr and serum starved for 6 h, and cells were immuno-stained with Rab8 (green) and Cep164 (red) Representative images are shown. Scale bar, 1 µm. (B) Cells with centrosomal Rab8 signal were counted (n≥100 per sample in two independent experiments). Error bars, SD. *p<0.05. (C) Mib1 E3 ligase activity antagonizes Rab8 recruitment to mother centriole. (D) Quantification of Rab8 recruitment to mother centriole in cells induced to ciliate in the presence of exogenous wild-type or catalytically inactive Mib1. (C) and (D) RPE1 cells were infected with control (Flag only), Flag-Mib1-WT or Flag-Mib1-C985S lentiviruses for 72 hr and serum starved for 6 hr. Cells were immuno-stained with antibodies against Rab8 (red) and Cep164+GT335 (green). N ≥ 100 per sample in two independent experiments. Error bars, SD. *p<0.05.
Additional files
-
Supplementary file 1
Statistical information.
- https://doi.org/10.7554/eLife.12950.019





