Transcription-coupled genetic instability marks acute lymphoblastic leukemia structural variation hotspots
Figures
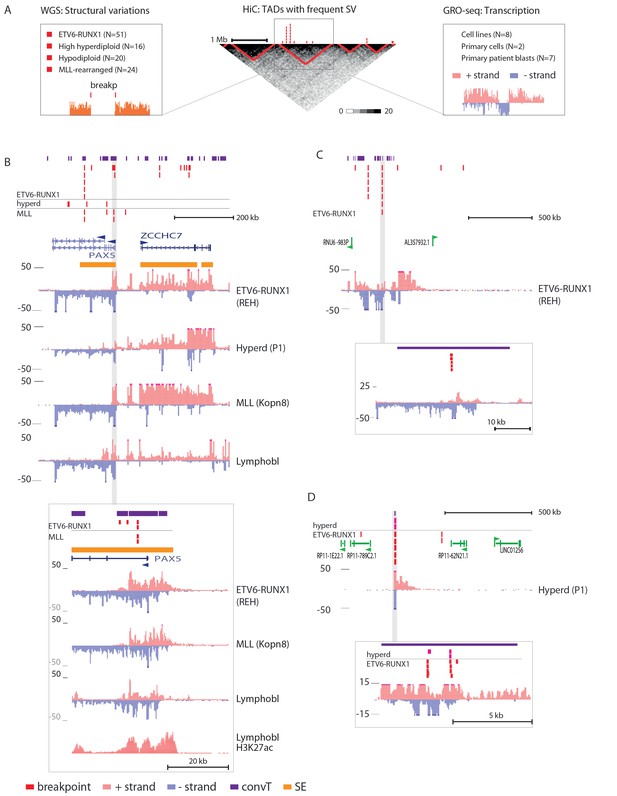
Integrative analysis of transcription and high-recurrence SV sites highlights novel transcribed regions.
(A) WGS data from the ETV6-RUNX1 (51 cases; Papaemmanuil et al., 2014), high hyperdiploid (16 cases; Paulsson et al., 2015), hypodiploid (20 cases; Holmfeldt et al., 2013) and MLL-rearranged (22 cases; Andersson et al., 2015) subtypes of precursor B-ALL was integrated with profiles of transcriptional activity assayed using GRO-seq from ALL patient and cell line samples (see also Figure 1—figure supplement 1 and Supplementary file 1). HiC data from B-lymphoid cells (Rao et al., 2014) was used to define TADs based on the HiC interaction frequency, shown as grey scale heatmap, in order to distinguish TADs with highest frequency of SV. (B) The PAX5 and ZCCHC7 loci are located in the TAD shown that has high SV frequency in hyperdiploid, ETV6-RUNX1- and MLL-fusion positive patients (4, 20 and 6 breakpoints, respectively, Figure 1—source data 1). The GRO-seq signal profiles from three pre-B-ALL cytogenetic subtypes and normal B-lymphoblastoid cells are displayed as indicated in the figure (see also Figure 1—figure supplement 4 and Figure 2—figure supplement 2). The y-axis shows the normalized read density (plus strand in red, minus strand in blue). convT regions regions are indicated in purple and leukemia breakpoints in red. The TSS region of PAX5 overlaps convT that co-localized with an intragenic SE (B-lymphoblastoid H3K27ac track is shown at the bottom). (C) A TAD with the same number of breakpoints (20) in ETV6-RUNX1 patients is shown with signal from REH cells (see also Figure 1—figure supplement 4). Genomic annotations include the location of GENCODE transcripts (in green). A strong transcription signal is visible that spans approximately 500 kb near the TAD boundary, lacking annotated transcripts. A zoom-in panel shows the most recurrent SV site. (D) The TAD visualized represents a genomic region that harbors most SV in HeH (see Figure 1—figure supplement 5 for the hypodiploid SV hotspot). The GRO-seq signal (track from patient 1) indicates a novel locus with abundant transcription in leukemic samples (refer to Figure 1—figure supplement 4 for all GRO-seq profiles). The highest recurrence of SV occurs at the convT overlapping mid-region (zoom-in panel), which has also two ETV6-RUNX1 breakpoints.
-
Figure 1—source data 1
Identified topologically associated domains.
The chromosomal coordinates (hg19) of TADs with breakpoints and all TADs as identified based on HiC data are reported. The number and frequency of breakpoints, together with% of domain spanned by convT are given for TADs with breakpoints. The highlighted rows correspond to TADs shown in Figure 1 and Figure 2—figure supplement 1. Notice the separate worksheets.
- https://doi.org/10.7554/eLife.13087.004
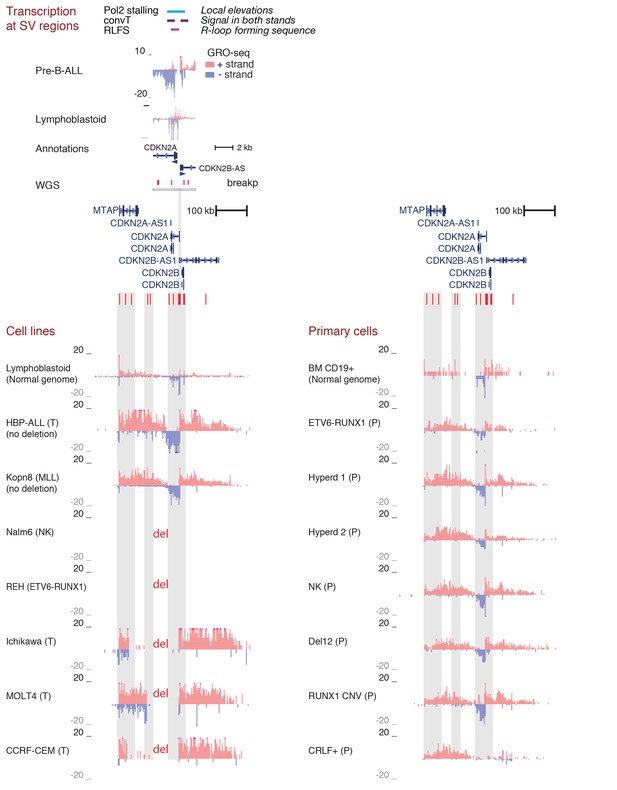
Transcriptional activity in leukemic cells from patients, cell lines and primary healthy B-lineage cells is captured in GRO-seq signals.
Transcriptional activity from the panel of GRO-seq samples is shown at the CDKN2A locus, representing a common deletion region. The closer view from the TSS region (above) illustrates the signal features of convT and Pol2 stalling that were computationally analyzed genome-wide (signal is shown from Kopn-8 and B-lymphoblastoid cells). SV present in a subset of the assayed cell lines can be distinguished based on the loss of signal across an extended region (indicated as 'del'). The cell lines with intact locus are shown first, followed by REH and Nalm6 cells that carry an extensive deletion and thus have no signal in the region shown, and three T-ALL cell lines with compact deleted regions. Primary patient data from diagnostic samples (right) can be examined in context of the cell lines (left) that represent complex genomes at relapse. B-lymphoblastoid and bone marrow CD19+ cells represent normal healthy cells. The Kopn-8 cell line carries an MLL rearrangement. The y-axis in the tracks shows the normalized read density and transcription from different strands is shown in tones of red (+ strand) and blue (- strand) for clarity. NK = normal karyotype, T = T-ALL, P=patient sample.
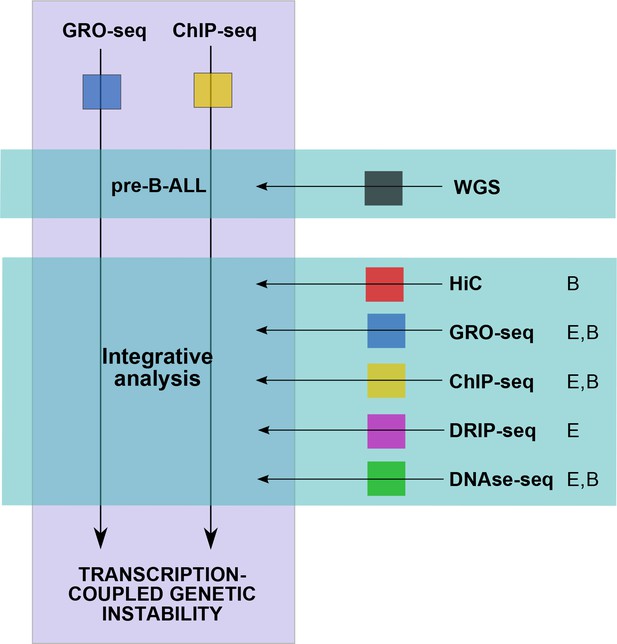
Summary of data used in the integrative analysis.
GRO-seq and ChIP-seq data were generated from pre-B-ALL cells (Supplementary file 1) for studying transcription-coupled mechanisms in context of leukemia SV. The SV data was retrieved from four published WGS studies (Holmfelt et al., 2013; Papaemmanuil et al., 2014; Andersson et al., 2015; Paulsson et al., 2015) and jointly analyzed with the signal profiles. To distinguish domains in the genome with high SV frequency, HiC data was retrieved from B-lymphoblastoid cells (Rao et al. 2014) and used to define TADs (Figure 1—source data 1). This initial analysis led to the identification of specific transcription signal features that occur frequently at breakpoints (convT and Pol2 stalling, Figure 2—source data 1). In order to characterize the properties of convT and Pol2 stalling sites genome-wide, additional analysis utilized further genomic profiles from B-lineage (B) and ES (E) cells (The ENCODE Project Consortium, 2012; Ginno et al., 2013). Inclusion of normal cell types allowed us to control for possible caveats in analyzing the signal from complex cancer genomes. In addition to the data shown, RSS and RLFS sequence motif analysis was used to distinguish DNA-encoded features.
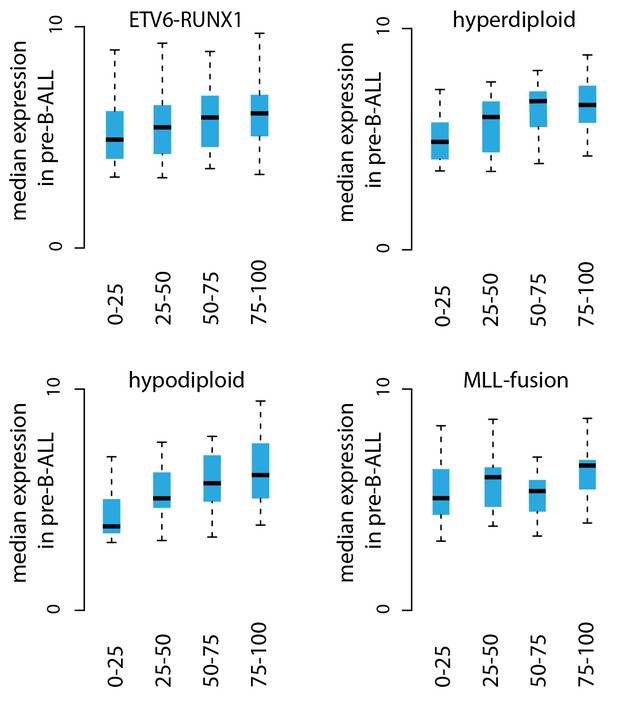
Transcriptional activity in TADs binned by breakpoint frequency.
The median transcription level (log2 signal) in pre-B-ALL patients (N = 1382) is summarized as boxplots from TADs divided into quartiles based on number of breakpoints per bp.
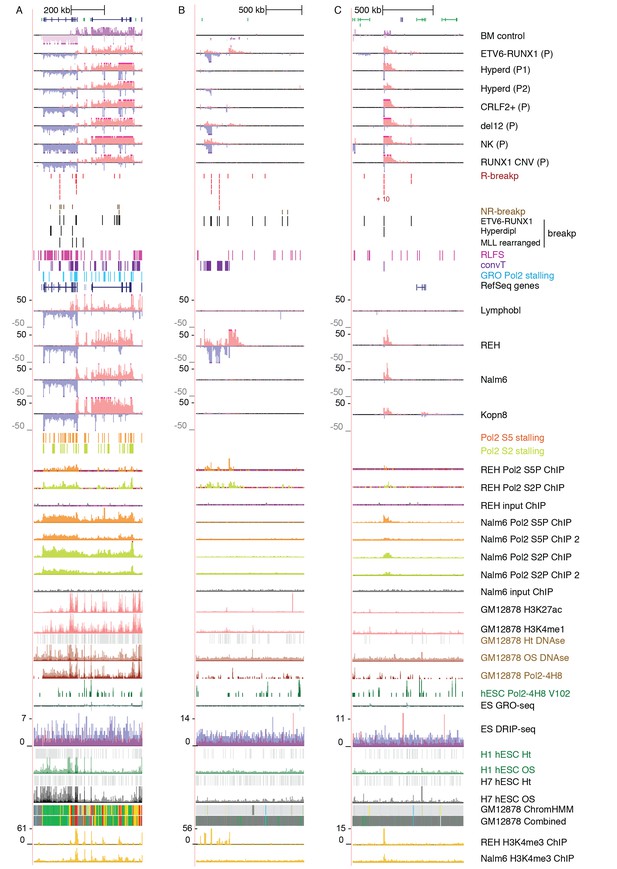
Data from all signal tracks for regions displayed in Figure 1.
The comprehensive set of signal and annotation tracks shown are organized as follows: Leukemia patient GRO-seq tracks are shown first, followed by leukemia breakpoint data. Next, vulnerable regions based on transcriptional features and cell line GRO-seq tracks are shown, followed by additional supporting tracks (Pol2 ChIP-seq, chromatin mark ChIP-seq and DNAse-seq from B-lineage and ES cells). Unless otherwise indicated, the y-axis in the tracks are -25:25 in GRO-seq, 0:25 in Nalm6 ChIP-seq. 2:10 in REH ChIP-seq, 0:50 in DNAse-seq OS, 0:0.5 in DNAse-seq DS, 0–100 in Layered H3K27ac (from GM12878) and 0–50 in Layered H3K4me1 (from GM12878). The cytogenetic subtype in GRO-seq samples is indicated with abbreviations (refer to Supplementary file 1) and P denotes primary patient samples. Bone marrow CD19+ cells from healthy donors and B-lymphoblastoid cells serve as controls. (A) Region shown corresponds to Figure 1A. Similarly, the two TADs harboring non-coding transcripts are shown in B and C. B. The novel transcripts are also detected in a patient with normal karyotype (NK) and another patient with deletion of chr12 (del12).
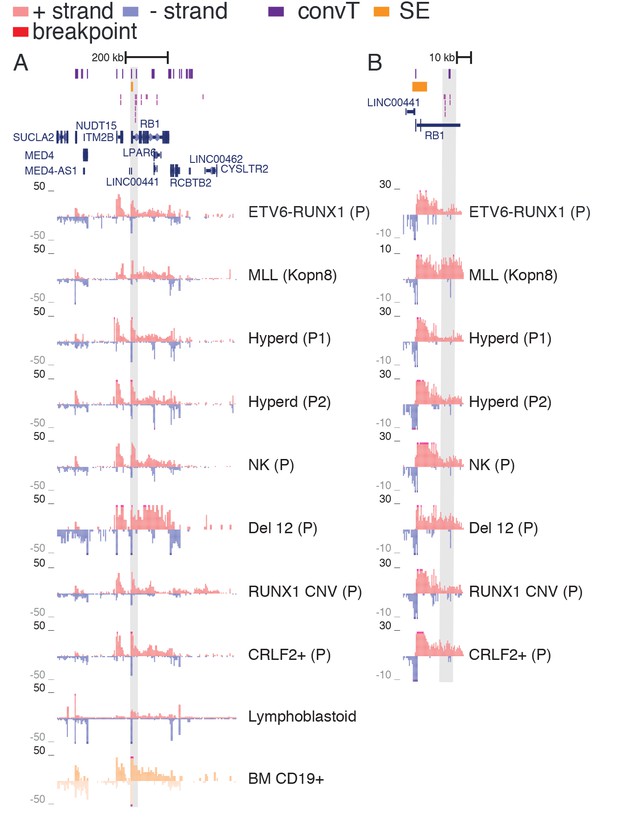
TAD with frequent SV in hypodiploid patients.
(A) The TAD visualized represents a genomic region that harbors most frequent SV in hypodiploid ALL. The GRO-seq signals (shown as in Figure 1) represent pre-B-ALL patients or cell lines with different cytogenetic subtypes. Notice that GRO-seq signal specifically for a hypodiploid case is not available. (B) The highest recurrence of SV occurs at the convT overlapping region downstream RB1 TSS.
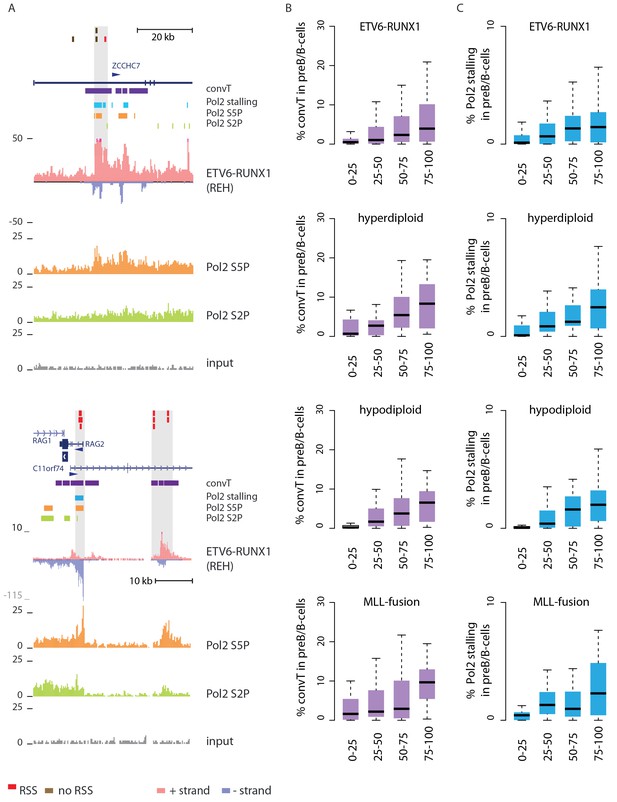
Convergent transcription and Pol2 stalling characterize genomic regions with high number of breakpoint events.
(A) The GRO-seq signal in the ETV6-RUNX1 positive REH cell line is shown to exemplify the co-occurrence of convT (in purple) and local elevation in GRO-seq signal (Pol2 stalling, in light blue) at both R- and NR-breakp (in red and brown, respectively) that reside within intronic (ZCCHC7), TSS (RAG2) or putative enhancer regions (RAG2). The elevated signal is also visible in Pol2 ChIP-seq signal (Pol2 S2P in green, Pol2 S5P in orange, input in grey). See also Figure 2—figure supplement 1. The percentage of TAD spanned by convT (in B) or Pol2 stalling (in C) in pre-B/B-lymphoid cells is summarized as boxplots from TADs divided into quartiles based on number of breakpoints per bp (see also Figure 1—figure supplement 3, Figure 2—figure supplement 3–6). The quartile ranges are for exclusive lower and inclusive upper value in the range, as indicated. Refer to Figure 2—source data 1 for statistical analysis.
-
Figure 2—source data 1
Identified convT and Pol2 stalling regions.
Regions with transcription from both strands (convT) or change points in the intragenic signal (Pol2 stalling sites) identified from B-lineage cells (REH, Nalm6 and Lymphoblastoid) or ES cells are listed with coordinates in the hg19 human genome. Coordinates of putative enhancers in B-lineage cells based on eRNA analysis (hg19 genome version) are shown in the second work sheet. The statistical analysis for genome-wide enrichment and the TAD analysis are included as separate worksheets, including also the results obtained based on ENCODE chromatin segmentation. Related to Figures 2–4.
- https://doi.org/10.7554/eLife.13087.011
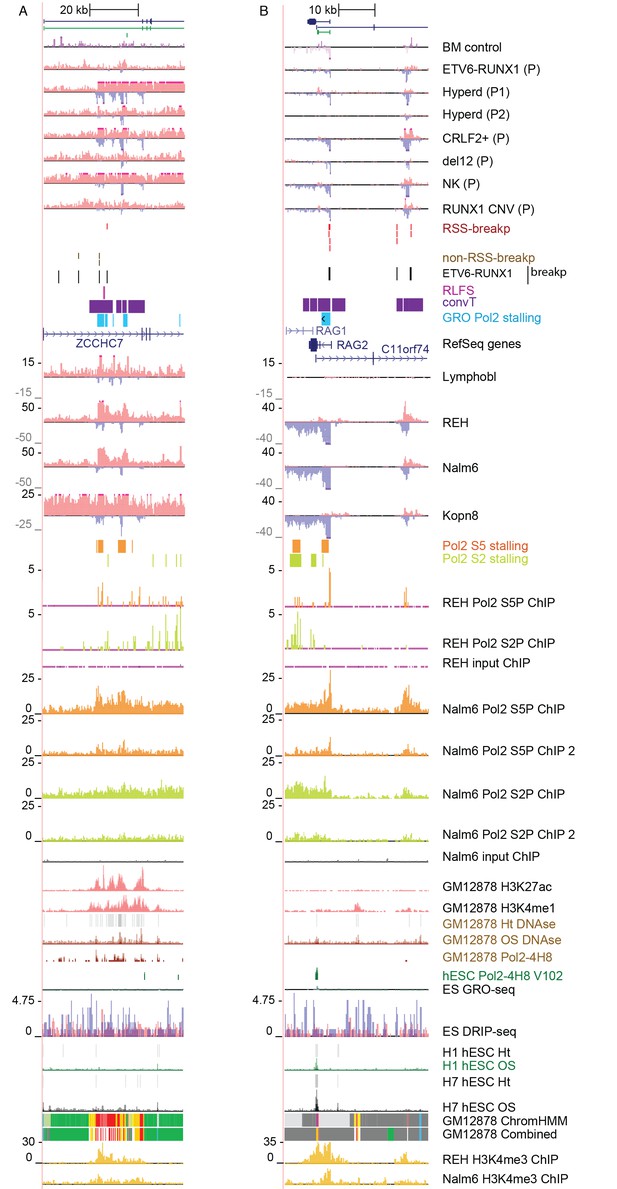
Data from all signal tracks for regions displayed in Figure 2.
The comprehensive set of signal and annotation tracks shown are organized as in Figure 1—figure supplement 4. The regions displayed correspond to those in Figure 2A. The ZCCHC7 intronic region is shown in A and the RAG2 locus in B.
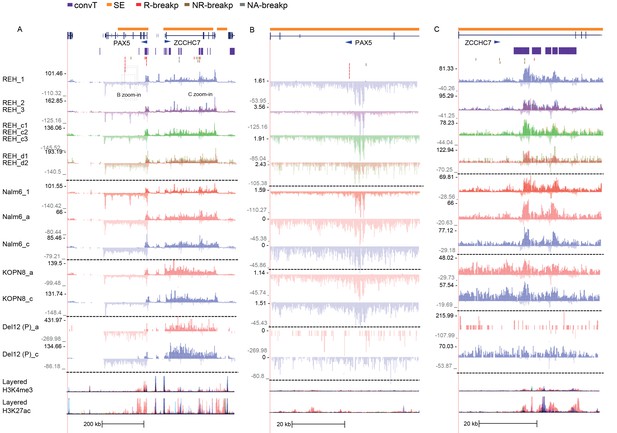
The GRO-seq signal from replicate samples generated from ALL cells displayed at the PAX5/ZCCHC7 locus.
A subset of GRO-seq samples were collected in multiple replicates in basal conditions from cells representing different genetic subtypes of ALL (refer to Supplementary file 1 for details). The separate tracks (shown in panels A–C) correspond to independent experiments and the overlaid signals show the signal profile from biological replicates collected within each experimental condition. (A) The PAX5/ZCCHC7 locus is shown (see also Figures 1 and 2), indicating the two regions that are displayed in more detail in panels B and C. The location of genes (in dark blue), SE (in yellow), convT regions (in purple) and breakpoints (R-breaks in red, NR-breaks in brown and unassigned in beige) are indicated above the signal tracks.
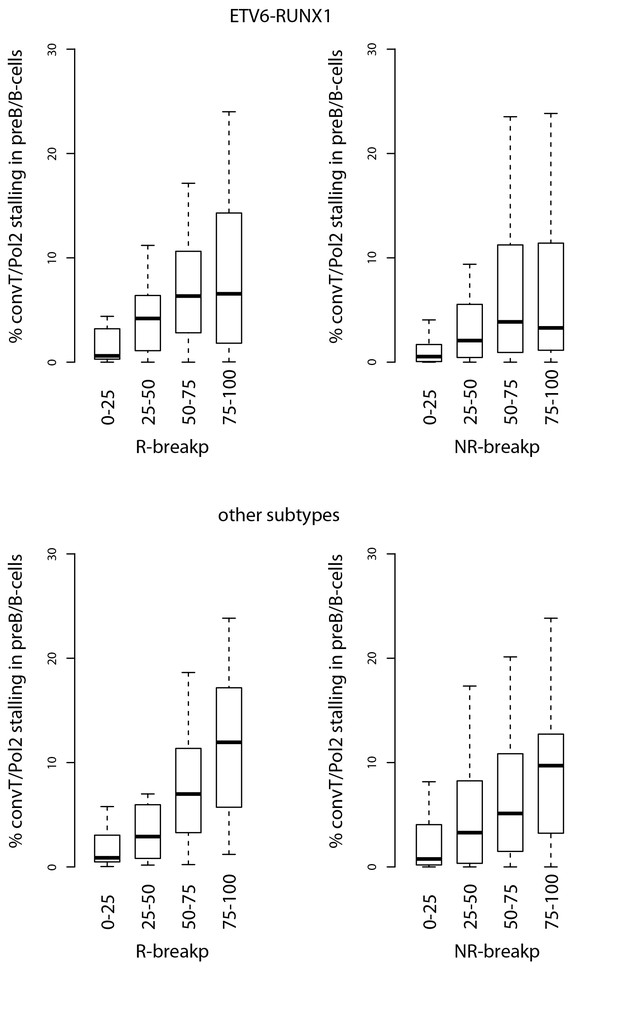
Signal feature span for TADs ordered separately by R-breakp or NR-breakp frequency.
The overlap percentage for convT and Pol2 stalling is shown as in Figure 2. The ETV6-RUNX1 data compared to other pre-B-ALL subtypes is displayed. The RSS status annotation is based on motifs identified from ETV6-RUNX1 breakpoints (see Materials and methods).
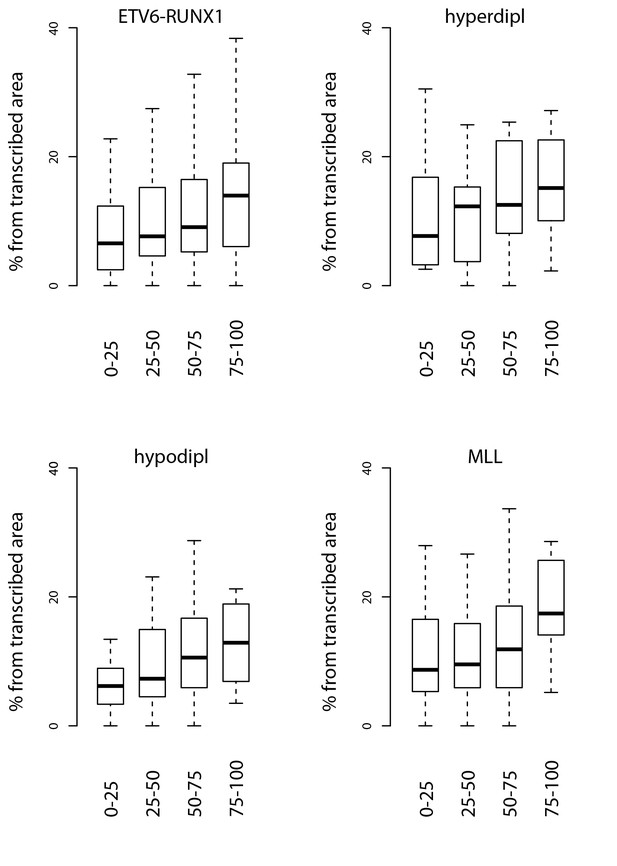
Signal feature span normalized by total transcribed area for TADs sorted by breakpoint frequency.
The percentage from total transcribed area that corresponds to convT or Pol2 stalling is shown for TAD quartiles based on the the four different pre-B-ALL subtype SV datasets.
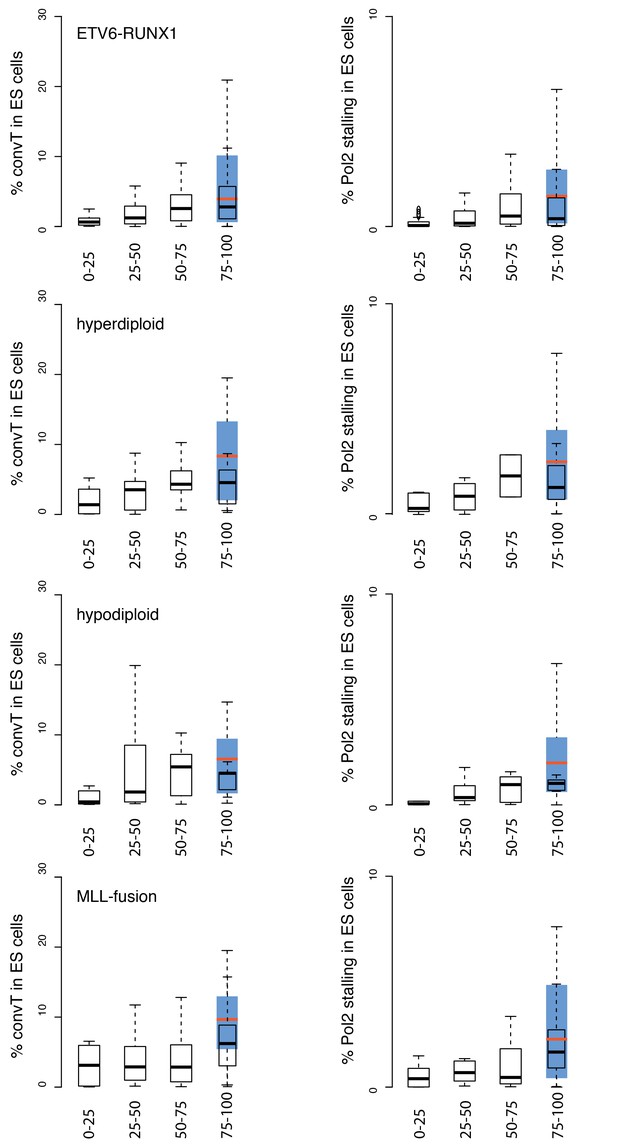
Overlap of TADs with convT in ES cells.
The percentage of TAD spanned by convT in ES cells is summarized as boxplots, as in Figure 2B and C. The relative difference to results presented in Figure 2 reflect cell type-specific transcriptional activity. To ease the comparison, the interquartile ranges for the overlap using relevant GRO-seq signals from pre-B/B-lymphoid cells are shown in blue (median plotted in red) for the highest SV frequency bin. The quartile ranges are for exclusive lower and inclusive upper value in the range, as indicated.
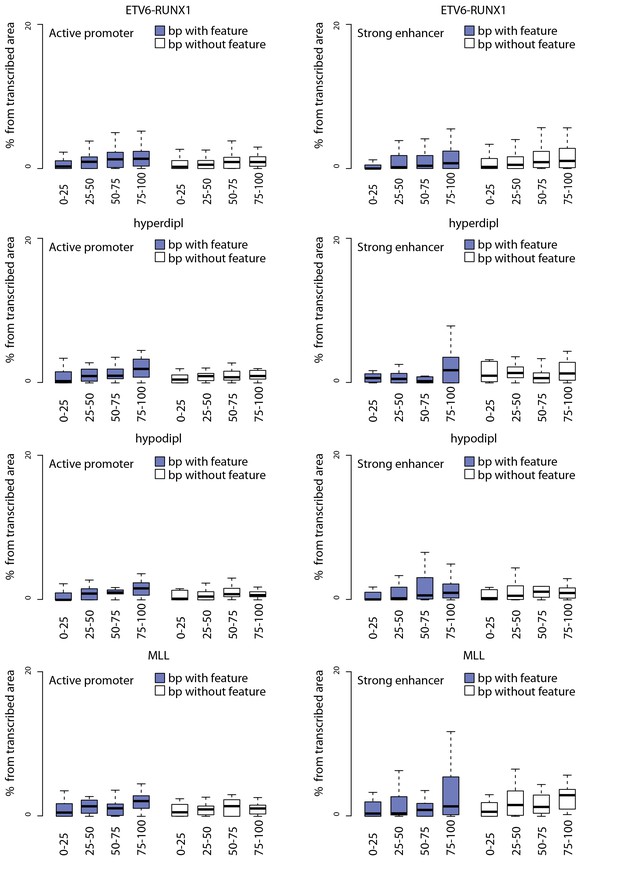
TAD analysis using promoter and enhancer chromatin segments stratified by convT and Pol2 stalling.
Contribution of convT and Pol2 stalling within active promoter and enhancer regions retrieved based on the chromatin segmentation of ENCODE B-lymphoblastoid cells is compared. Subregions within active promoters/enhancers that overlap convT/Pol2 stalling (in colored boxplots) or lack these features (no color) were used in the analysis comparing TAD quartiles. The data are normalized by total transcribed area as in Figure 2—figure supplement 4.
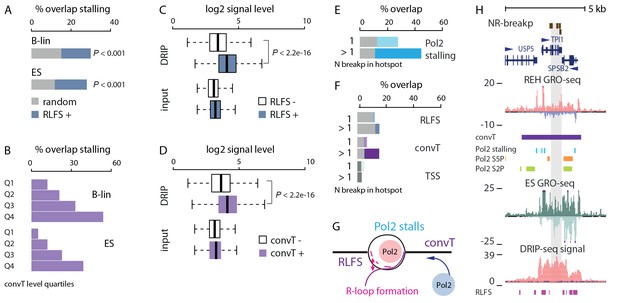
Indication of transcription-coupled genetic instability at leukemia SV hotspots lacking RSS motifs.
(A) Overlap between RLFS motif harboring intragenic regions and detected Pol2 stalling sites in B-lineage and ES cells. The high overlap of RLFS-positive regions is statistically significant compared to random regions (empirical P is indicated for 30% and 28% overlaps, respectively). (B) Overlap of detected Pol2 stalling sites also increases based on the strength of antisense signal level for B-lineage and ES cell convT regions divided into quartiles. (C) The influence of RLFS at TSS on ES cell DRIP-seq signal level is shown (Wilcoxon rank sum test P is indicated). Input signal levels are shown as control. (D) ES cell DRIP-seq signal is plotted similarly as in C, from convT-positive and -negative TSS regions. The DRIP-signal is higher in convT-positive TSS (Wilcoxon rank sum test P is indicated, TSS with convT N = 11774, TSS without convT N = 12092, refer to Figure 3—source data 2 for statistical analysis based on separate DRIP-seq replicates). (E) The percentages of breakpoint regions with no RSS motifs overlapping intragenic Pol2 stalling sites found in B-lineage cells are shown as barplots. The mean overlap observed in random sampling is indicated in grey bars (further statistical analysis is presented in Supplementary file 3). Categories with increasing cut-off for recurrence (1: non-recurrent in dim color, >1 and above: recurrent in darker color) were tested. (F) Overlap with RLFS, convT and annotated TSS is shown, as in E, for ETV6-RUNX1 NR-breakp (see also Supplementary file 3). (G) A schematic model illustrating how transcription from both strands (convT) or RLFS can locally arrest the Pol2 complex leading to recruitment of DNA damage-sensing complexes to R-loops, such as AID or BRCA (Alt et al., 2013, Hatchi et al., 2015), in an RSS-independent manner. (H) NR-breakp hotspot with the highest recurrence (TPI1 locus) is shown. DRIP-seq signal (shown in tones of red overlaid with input control signal in blue), and RLFS motifs indicated as a magenta bar track represent two levels of independent data that were integrated with GRO-seq data (signal from REH and ES cells is shown) to characterize properties of convT and Pol2 stalling regions. The breakpoint data (NR-breakp in brown) and detected convT (in purple) and Pol2 stalling in B-lineage cells (in blue) are shown. At the the recurrent breakpoint sites antisense transcription of neighboring gene (SPSB2 primary transcript) leads to a broad convT region, as indicated in the figure. Elevated DRIP-signal indicates formation of DNA-RNA hybrids (see also Figure 3—figure supplement 3).
-
Figure 3—source data 1
Breakpoint clustering to regions.
ETV6-RUNX1 breakpoint data used in the analysis was divided into three categories based on evidence for RSS-guided RAG targeting to the region (RSS-motifs). To analyze recurrence, breakpoint events within 1-kb distance were stitched together. The resulting genomic region coordinates (hg19) and the number of breakpoints contained within them are reported sorted by breakpoint count. Statistical analysis of feature overlap based on binomial and hypergeometric distribution is summarized in the following worksheet. Coordinates and statistics for all pre-B-ALL breakpoint regions are listed in the last worksheet. Notice the separate worksheets.
- https://doi.org/10.7554/eLife.13087.019
-
Figure 3—source data 2
Statistical analysis of separate DRIP-seq and DNAse-seq replicates.
- https://doi.org/10.7554/eLife.13087.020
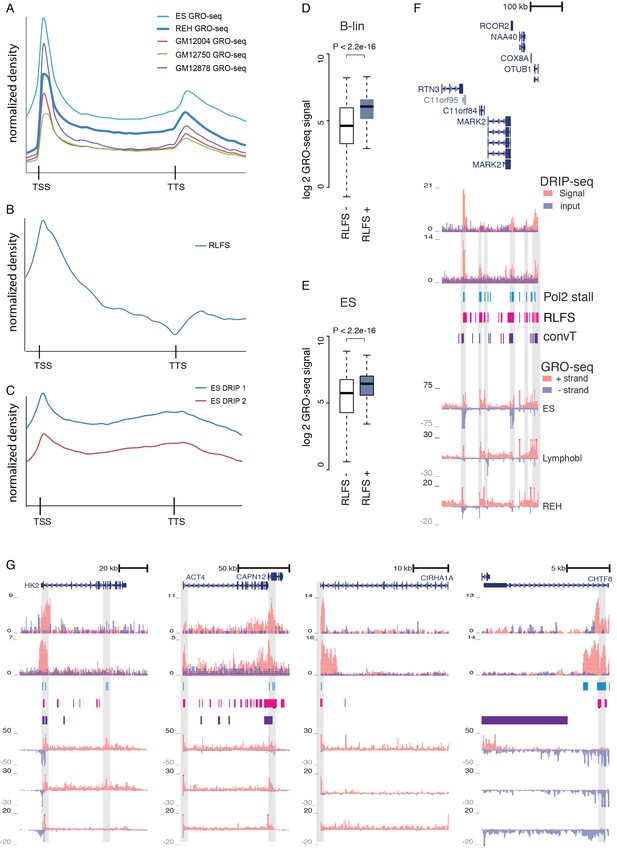
GRO-seq, RLFS and DRIP-seq signal profiles across genes.
(A–C) Signal levels summarized across length-normalized transcribed gene loci: (A) GRO-seq in ES, REH and B-lymphoblastoid cells, (B) RLFS motif counts, (C) DRIP-seq signal in ES cells. The y-axis has an arbitrary scale normalized across all data points and the x-axis indicates the start and end of transcripts. The GRO-seq signal density across genes (in A) shown allows distinguishing signal increases that are indicative of Pol2 stalling. The TSS and TTS regions harbor sequence elements that favor formation of R-loops. (D) GRO-seq signal (B-lineage cells) compared between TSS with or without RLFS motifs is shown as box plots. The signal distributions differ significantly (Wilcoxon rank sum test P is indicated, TSS with RLFS N = 15646, TSS without RLFS N = 8220). (E) GRO-seq signal (ES cells) compared between TSS with or without RLFS motifs as in (D, F) An example genome region with high density of RLFS. DRIP-seq signal is shown in tones of red overlaid with input control signal in blue, RLFS motifs are indicated as a magenta bar track, GRO-seq overlaid signal from replicate samples is shown with + strand in red and - strand in blue. (G) Experimentally verified R-loop rich regions (Ginno et al., 2013) are shown as in F. The DRIP-seq signal is elevated frequently at TSS and TTS regions as shown at HK2, ACTN4, CIRH1A and CHTF8 loci. Correspondence between RLFS density, GRO- and DRIP-seq signal levels and detected signal features (convT and Pol2 stalling) can be further examined based on tracks shown.
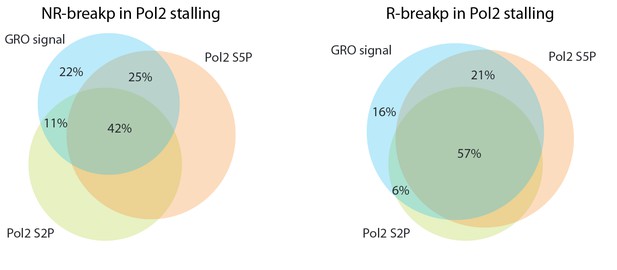
Venn diagrams comparing SV within Pol2 stalling regions based on GRO- and ChIP-seq profiles.
The Pol2 stalling regions overlapping SV detected using GRO-seq, Pol2 S2P and Pol2 S5P ChIP-seq are compared in the Venn diagrams shown. Data for NR- and R-breakp is shown separately.
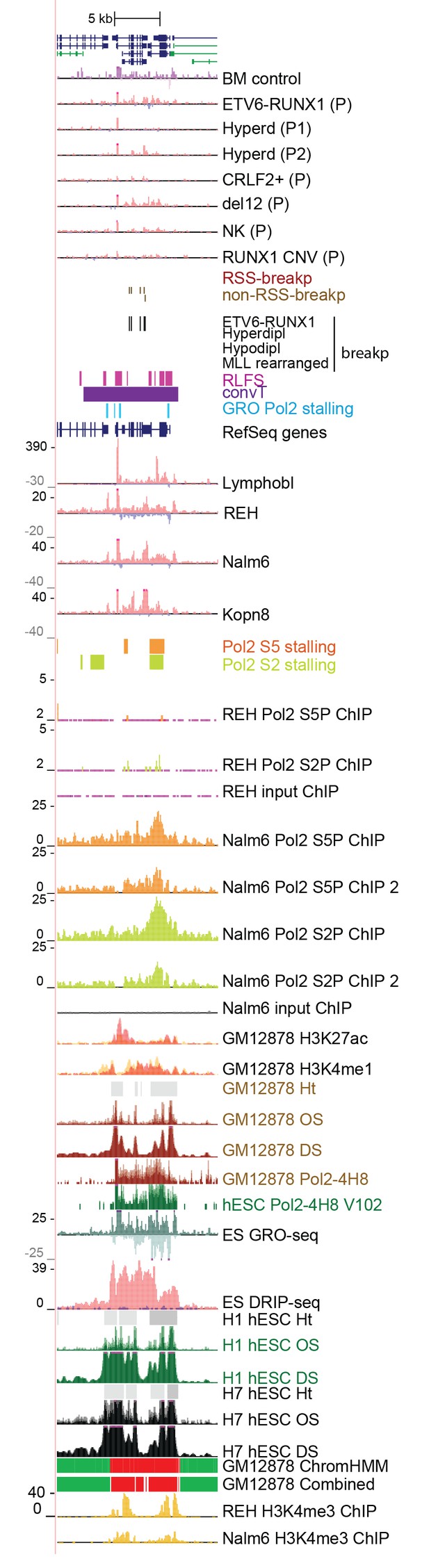
Data from all signal tracks for regions displayed in Figure 3.
The comprehensive set of signal and annotation tracks shown are organized as in Figure 1—figure supplement 4. The region displayed correspond to the TPI1 locus shown in Figure 3H.
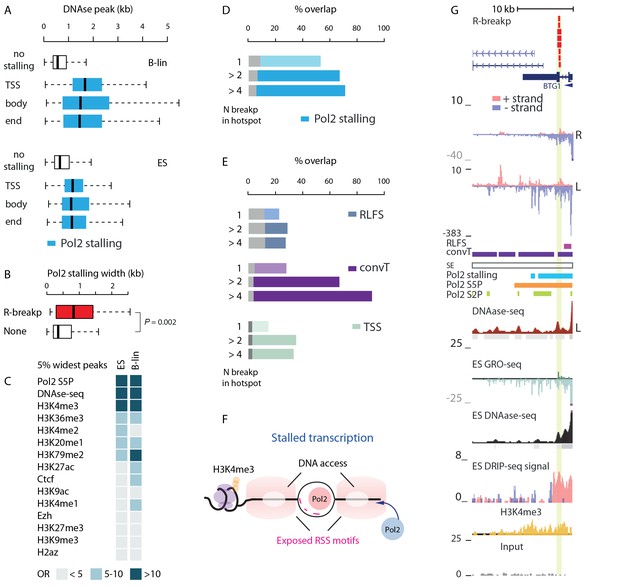
SV with RSS motifs localize to Pol2 stalling regions with broad open chromatin regions.
(A) DNA access based on DNAse-seq peak width (GM12878 or H1 ES from ENCODE) is compared between regions with no Pol2 stalling (no color) and overlapping Pol2 stalling (light blue, cell-specific Pol2 stalling coordinates are listed in Figure 2—source data 1) at TSS, body and end region of transcripts (refer to Figure 3—source data 2 for statistical analysis based on separate DNAse-seq replicates). (B) The TSS stalling width is compared between TSS harboring R-breakp and TSS with no breakpoints (Wilcoxon rank sum test P is indicated, TSS with R-breakp N = 38, TSS without breakpoints N = 11957, 95% CI for size difference 67–491 bp). (C) The 5% widest Pol2 stalling regions were overlapped with similarly defined widest peaks in different ChIP- and DNAse-seq data (refer to Figure 4—source data 1 for details and all statistics). The odds-ratio (OR) for the overlap is visualized in color from discrete categories (<5; 5–10; >10, with darker color tones indicating higher OR). Pol2 S5P, DNAse-seq and H3K4me3 peaks had highest OR based on both B-lineage and ES cell data. D and E: The percentages of R-breakp overlapping Pol2 stalling (as in Figure 3E) or RLFS, convT and annotated TSS (as in Figure 3F) are shown as barplots, respectively. Overall, the recurrence was higher compared to NR-breakp and therefore two categories for recurrent R-breakp are shown (>2; >4). The overlap with convT reaches 91% at highly recurrent R-breakp hotspots (source data can be found in Figure 2—source data 1, S6 and statistics for genes binned by their transcription level in Supplementary file 3). (F) A schematic model illustrating how the transcriptional features may lead to the recruitment of RAG1 and RAG2 based on RSS-motif recognition and chromatin. Pol2 stalling associated with DNA accessibility and wide deposition of the H3K4me3 mark. (G) R-breakp hotspot with the highest recurrence (BTG1 locus) is shown. B-lymphoblastoid and ES cell tracks from DNAse-seq and H3K4me3 from pre-B-ALL cells (Nalm6) represent signals with highest overlap to wide Pol2 stalling (other tracks as in Figure 3H, see also Figure 4—figure supplement 1).
-
Figure 4—source data 1
Overlap of wide Pol2 stalling regions with unusually wide peaks representing other chromatin features.
The table summarizes the highest observed odds ratios in the Fisher test for the overlap between top 5% widest chromatin features and 5% of widest Pol2 stalling regions. Empirical p-values are reported together with the Fisher test values separately for in ES and B-lineage cells. Data for the different replicate experiments are shown as a separate work sheet.
- https://doi.org/10.7554/eLife.13087.025
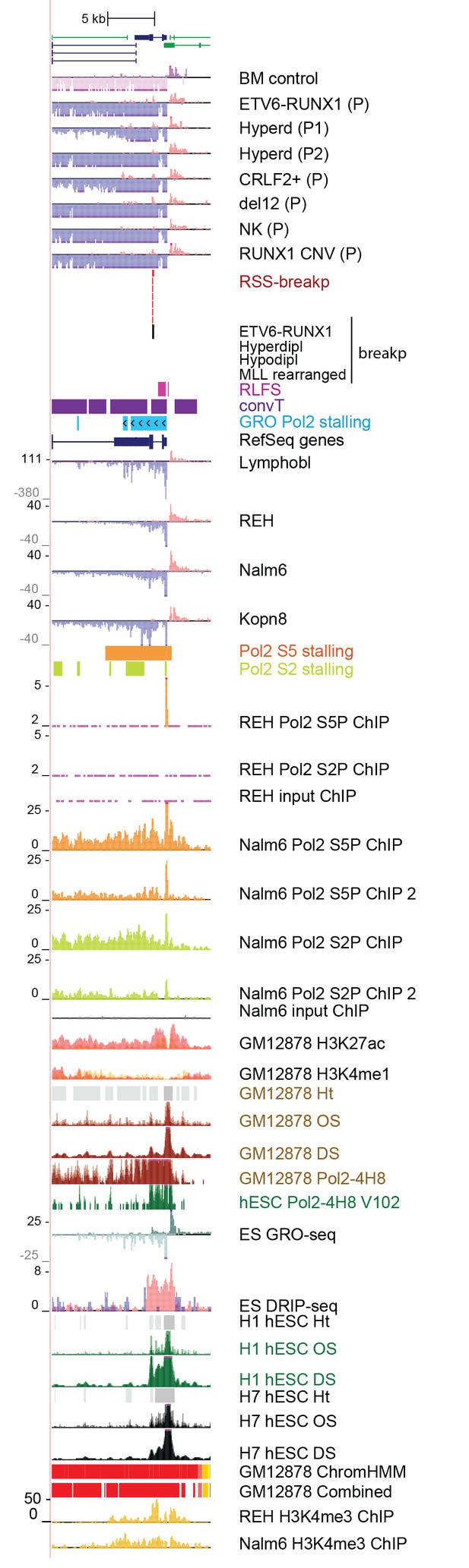
Data from all signal tracks for regions displayed in Figure 4.
The comprehensive set of signal and annotation tracks shown are organized as in Figure 1—figure supplement 4. The regions displayed correspond to the BTG1 locus shown in Figure 4G. The ZCCHC7 intronic region is shown in A and the RAG2 locus in B.
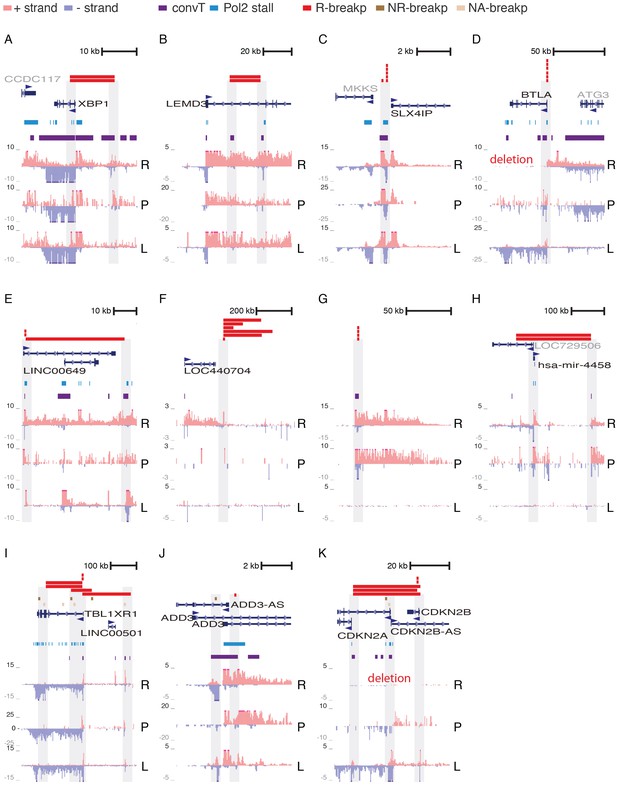
GRO-seq signal profile at multiple clustered deletion regions.
Further examples of R-breakp regions, representing deletion clusters defined in (Papaemmanuil et al., 2014) (see Supplementary file 2 listing genomic coordinates shown). The transcription signal at breakpoint ends is highlighted in each panel. Those breakpoints that are contained within the region are shown as wider colored bars. For breakpoints that end outside the region only one end is indicated (in panel G the short bars correspond to the full deletion regions that are very local). The regions are sorted in the following manner: coding gene loci that harbor R-breaks, similar non-coding gene loci, gene loci with R- and NR-breaks (including a few with no RSS-motif status, denoted NA-breakp). convT regions with short 1–2 kb transcripts on the opposite strand, typical of enhancer RNAs, overlap several R-breakp. To exemplify, in panel A, a distal upstream enhancer (and possibly an intragenic enhancer) nearby XBP1 co-localizes with breakpoints while in B, the deletion occurs at short distance involving two intragenic enhancers of LEMD3. Notice that the cluster region shown in D and K harbor deletions in the REH cell line. In both cases the breakpoints are flanked by convT based on B-lymphoblastoid cell data. The TBL1XR1 locus (in I) harbors most frequent NR-breakp that overlaps with elevated GRO-seq signal assigned as Pol2 stalling regions. R: REH (ETV6-RUNX1 fusion), P: ALL patient (ETV6-RUNX1 fusion), L: Lymphoblastoid cell line (normal karyotype).
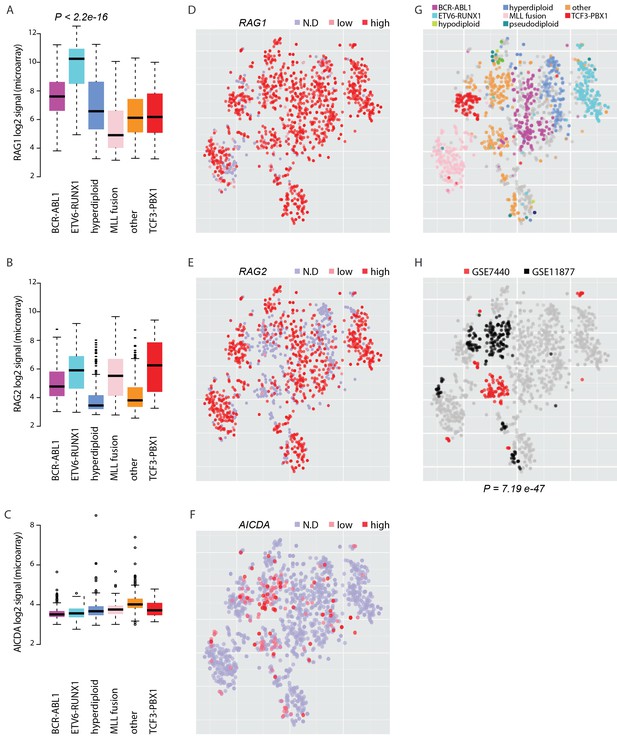
Expression of AID and RAG across molecular subtypes of leukemia.
The log2 expression signal is summarized as boxplots for (A) RAG1 (B) RAG2 and (C) AICDA across the pre-B-ALL subtypes (N = 153 BCR-ABL1, N = 153 ETV6-RUNX1, N = 151 hyperdiploid, N = 198 MLL rearrangement, N = 267 other, N = 82 TCF3-PBX1). Wilcoxon rank sum test p-value is indicated for differential RAG1 expression in the ETV6-RUNX1 subtype (N = 153, patients with cytogenetic subtype information N = 1008) (in A). (D–F) Alternative representation of discrete expression states for RAG1, RAG2, and AICDA, respectively (red: high, pink: low, grey: not detected). The data points shown as a t-SNE map correspond to the full set of pre-B-ALL patient samples (N = 1382) (see also Figure 5—source data 1). Their relative positions are defined by the transcriptome similarity. The sample groups can be compared to annotated cytogenetic types, as colored on the same map in (G, H). The location of high-risk samples (N=295) from two independent studies is indicated in color on the same map (COG studies GSE7740 in red and GSE11877 in black, see also Supplementary file 5). Hypergeometric test p-value is indicated for enrichment of detected AICDA expression in the high risk studies (N = 112, refer to Supplementary file 5 for population statistics).
-
Figure 5—source data 1
pre-B-ALL transcriptome samples.
Sample identifiers of pre-B-ALL transcriptomes analyzed and their coordinates on the dimensionality reduction plot.
- https://doi.org/10.7554/eLife.13087.029
Additional files
-
Supplementary file 1
GRO-seq sample summary.
Description of the patient and cell line GRO-seq samples used in the analysis, including the cell culture conditions, replicate information and the total number of pooled sequencing reads obtained after quality filtering and alignment. A more detailed table for cultured samples with replicate information and accession codes is provided at the bottom. Sample accession codes for already published and re-analyzed GRO-seq data, and additional GRO-seq data displayed in Figure 1—figure supplement 1 are listed in worksheet 2.
- https://doi.org/10.7554/eLife.13087.030
-
Supplementary file 2
Genomic coordinates for regions displayed.
The coordinates of example gene regions displayed in the main and supplementary figures are listed (hg19 human genome version).
- https://doi.org/10.7554/eLife.13087.031
-
Supplementary file 3
Breakpoint hotspot analysis for genes binned by the transcription level.
- https://doi.org/10.7554/eLife.13087.032
-
Supplementary file 4
Intragenic recurrent SV in ETV6-RUNX1 patients with overlap to vulnerable regions.
The patient and region identifiers for recurrent intragenic SV in ETV6-RUNX1 patients are listed, reporting separately those co-localized with Pol2 stalling or convT regions.
- https://doi.org/10.7554/eLife.13087.033
-
Supplementary file 5
Clinical data for patients with high AICDA expression.
Study description, sample identifier, cytogenetic group, age and dataset identifier are listed for the patients within high AICDA expression level. Statistical analysis testing enrichment of detected AICDA expression in high risk studies is summarized in worksheet 2.
- https://doi.org/10.7554/eLife.13087.034
-
Supplementary file 6
Custom blacklisted genomic regions.
Blacklisted regions discarded from the analysis that were deemed to represent low-mappability, rRNA and snoRNA loci based on GRO-seq signal. Coordinates refer to the hg19 human genome version.
- https://doi.org/10.7554/eLife.13087.035





