Evidence of human infection by a new mammarenavirus endemic to Southeastern Asia
Figures
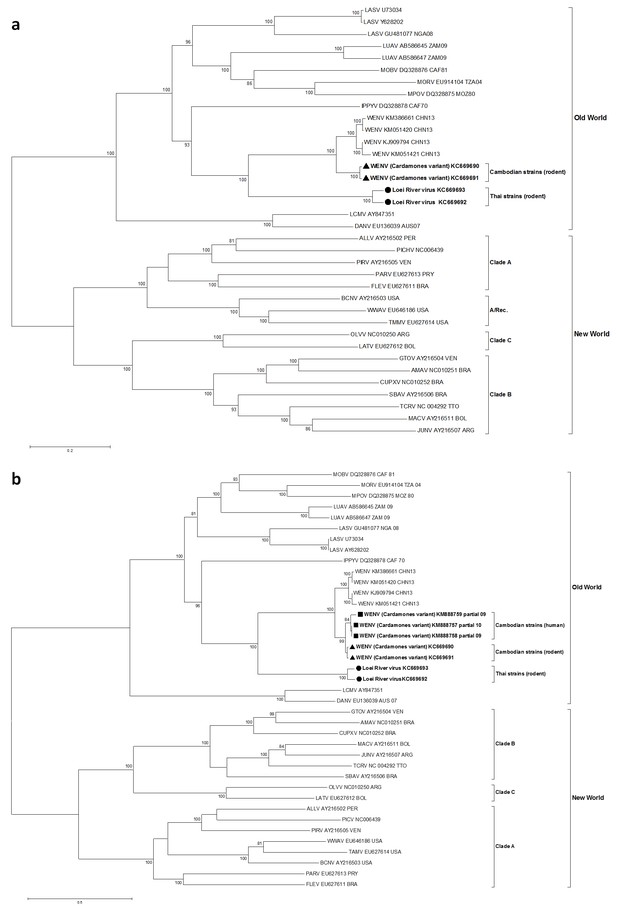
Maximum likelihood phylogenetic tree of novel arenavirus isolates and other representative arenaviruses for a, the complete ORF of the L gene with sequences from rodents only, b, partial L sequences including sequences from rodents and patients.
Cambodian strains detected in rodent (triangle), human (square) and Thai strains detected in rodent (circle) are in bold. Clade A, B and C are three evolutionary lineages of New World arenaviruses within the Tacaribe complex. A/Rec denotes the recombinant clade including the three Northern American viruses. The virus names are in abbreviation according to Radoshitzky et al. (2015).
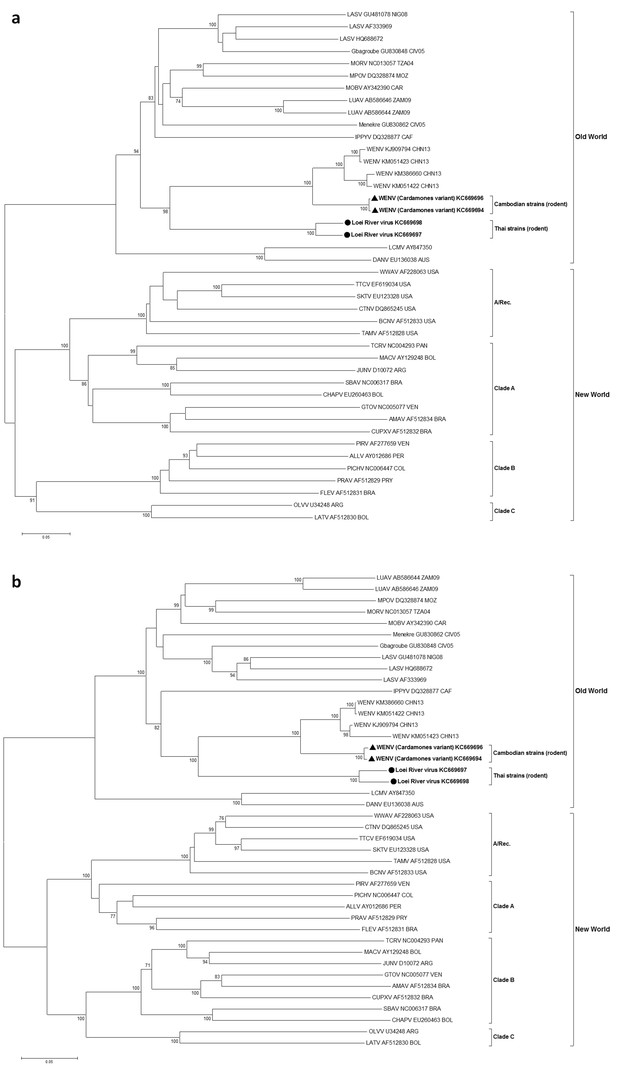
Maximum likelihood phylogenetic tree of novel arenavirus isolates and other representative arenaviruses for a, the complete ORF of GPC gene and b, complete ORF of NP gene.
Cambodian strains (triangle) and Thai strains (circle) detected in rodents are in bold. Clade A, B and C are three evolutionary lineages of New World arenaviruses within the Tacaribe complex. A/Rec denotes the recombinant clade including Northern American viruses. The virus names are in abbreviation according to Radoshitzky et al. (2015).
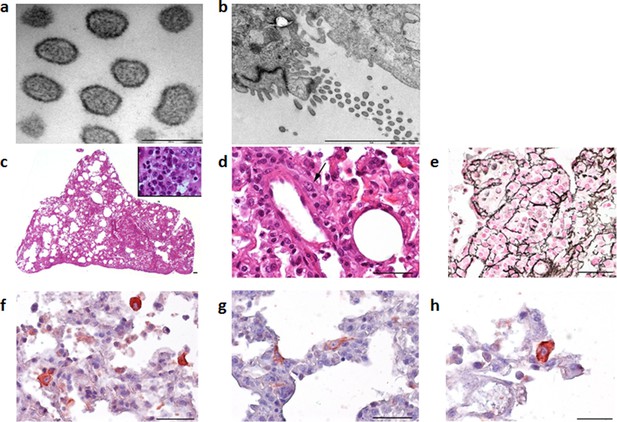
Electron microscope images of the Cardamones variant of Wēnzhōu virus in the rat lung tissues, a (scale: 200 nm) & b (scale: 2 µm).
Histopathological examination of the lungs revealed only severe diffuse pneumonia, with lesions associated with acute exudative inflammation characterised by foci of consolidation surrounded by extremely rare aerated parenchyma, c, vascular congestion, acute bronchiolitis and diffuse leukocytic infiltration with lymphocytes, polymorphonuclear and macrophages (suppurative exudate in the lumen and parietal inflammation), c and d. Reticulin staining demonstrated the severe destruction of lung parenchyma, e. Chromogenic immunohistochemistry identified inflammatory foci with numerous positive cells, f, primarily inflammatory cells including macrophages, but in the more preserved parenchyma and aerated parenchyma some epithelial (pneumocytes), g, and alveolar macrophages, h, were also clearly stained.
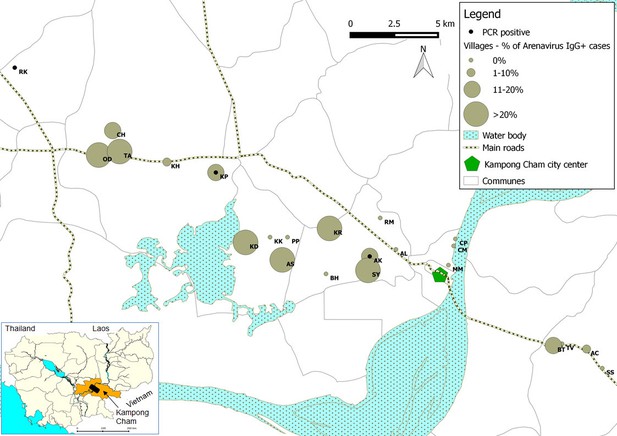
Representation map of the percentage (%) of patients who tested positive by anti-arenavirus IgG ELISA and of 6 patients who tested positive by L gene RT-PCR in the villages from Kampong Cham province.
Village names: AC=Andoung Chea, AK=Ampil Kraom, AL=Ampil Leu, AS=Andoung Svay, BH=Banteay Thma, BT=Boeng Tras, CH=Chachak, CM=Chong Thnal Muoy, CP=Chong Thnal Pir, KD=Kdei Boeng, KH=Kakaoh, KK=Kouk Kream, KP=Krasang Pul, KR=Krala, MM=Memay, OD=Ou Da, PP=Prey Phdau, RK=Roung Kou, RM=Romeas, SS=Srae Siem, SY=Sya, TA=Tuol Ampil and TV=Tuol Vihear.

Positions of primers used in diagnostic PCR assays.
The positions of the primers used for qRT-PCR are included in their names and these positions are based on the sequence of the L gene of Cardamones variant of Wēnzhōu virus. The positions of the primers used in the nested RT-PCR originate from the article of Vieth et al. (Vieth et al., 2007).
Tables
Nucleotide and amino acid sequence identities (%) between Cambodian and Thai isolates and selected other arenaviruses.
| Isolates | Segment or ORF | nt/ aa | Cambodian isolates | Thai isolates | Wēnzhōu | Lassa | Ippy | Mopeia | LCMV | Junín | Luna | Morogoro |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cambodian isolates | L segment | nt | 98.9 | 69.2-69.4 | 87.5-88.6 | 55.8-56.4 | 57.7-57.8 | 60.8 | 56.9 | 50.2-50.4 | 61.1-61.4 | 60.9-61.0 |
| L ORF | nt | 99.3 | 67.3-67.5 | 88.0-89.0 | 59.7-60.6 | 60.4-60.5 | 59.2-59.4 | 55.6 | 50.6-50.9 | 59.6-60.9 | 59.6 | |
| aa | 99.6 | 69.2-69.4 | 92.2-94.8 | 55.5-56-4 | 57.7-57.8 | 56.6-57.1 | 48.5 | 37.9-38.0 | 55.8-55.9 | 55.6 | ||
| Z ORF | nt | 99.5 | 73.4-74.5 | 837.-87.9 | 66.8-67.4 | 63.6 | 62.5 | 57.6-58.2 | 54.9-55.4 | 69.0-70.1 | 64.7-65.2 | |
| Aa | 98.8 | 79.2 | 89.4-93.9 | 70.1-75.3 | 70.1 | 64.9 | 59.4 | 40.3 | 63.6-64.9 | 61.0 | ||
| S segment | nt | 99.5 | 71.7-72.1 | 87.5-89.8 | 61.7-68.1 | 66.6-66.8 | 66.6-67.0 | 61.6-61.8 | 54.6-54.8 | 67.6-68.2 | 66.7-66.9 | |
| NP ORF | nt | 99.3 | 73.1-74.4 | 86.6-90.0 | 67.1-68.2 | 68.3-68.4 | 67.6-67.9 | 62.9-63.2 | 55.1-55.6 | 69.0-70.3 | 68.7 | |
| aa | 99.8 | 82.9-84.2 | 87.3-96.5 | 72.2-73.8 | 74.4-74.6 | 73.5-73.8 | 64.0 | 51.9-52.5 | 73.3-74.0 | 74.0-74.6 | ||
| GPC | nt | 99.7 | 69.1-70.0 | 88.6-89.7 | 67.3-68.6 | 64.7-65.7 | 65.3-65.7 | 61.2-61.4 | 53.4-53.6 | 66.3-66.6 | 49.9-65.2 | |
| aa | 99.8 | 79.5-81.1 | 95.5-96.4 | 74.2-76.2 | 69.5-71.3 | 71.5-72.8 | 57.2-57.5 | 48.2-43.0 | 72.6-74.4 | 73.1-74.4 | ||
| Thai virus | L segment | nt | 68-68.1 | 94.6 | 66.6-67.5 | 60.7-61.8 | 61.3-61.5 | 61.1-61.4 | 57.1 | 50.2-50.5 | 60.7-61.4 | 61.4-61.5 |
| L ORF | nt | 67.3-67.5 | 95.1 | 67.9-68.8 | 59.6-59.9 | 59.9-60.4 | 61.6-62.1 | 55.7-56.2 | 49.9-50.8 | 60.4-61.6 | 59.9-60.2 | |
| aa | 69.2-69.4 | 96.7 | 69.6-70.7 | 55.7-56.4 | 56.5 | 56.6-57.1 | 49.0-49.4 | 37.1-37.5 | 55.9-56.3 | 56.5-56.7 | ||
| Z ORF | nt | 73.4-74.5 | 95.4 | 69.4-75.0 | 65.2-66.8 | 64.1-65.8 | 69.0-69.6 | 58.7-59.2 | 54.3-56.0 | 65.8-72.8 | 66.3-66.8 | |
| aa | 79.2 | 98.5 | 73.1-74.6 | 67.5-71.4 | 68.8 | 70.1 | 58.4 | 42.9-44.2 | 70.1-71.4 | 66.2 | ||
| S segment | nt | 71.7-72.1 | 94.4 | 71.0-72.2 | 65.8-67.3 | 66.1-66.8 | 66.2-67 | 61.7-62.6 | 53.9-55.0 | 65.2-68.1 | 66.1-67.9 | |
| NP ORF | nt | 73.1-74.4 | 94.6 | 72.2-74.1 | 65.2-67.8 | 66.6-67.3 | 66.9-67.7 | 62.4-63.2 | 54.1-54.4 | 66.1-68.0 | 67.3-68.9 | |
| aa | 82.9-84.2 | 98.1 | 78.3-87.2 | 73.3-74.6 | 72.0-72.9 | 73.8-74.4 | 64.2-64.6 | 49.9-51.0 | 72.0-73.5 | 74-75.5 | ||
| GPC | nt | 69.1-70 | 94.1 | 69.2-69.9 | 66.0-67.6 | 65.0-66.1 | 66.4-67 | 61.5-62.6 | 53.9-56.0 | 65.8-69.2 | 66.1-67.5 | |
| aa | 79.5-81.1 | 97.5 | 80.1-81.1 | 73.5-75.7 | 71.5-72.4 | 71.0-74.2 | 59.7-61.0 | 43.0-43.9 | 74.2-74.8 | 73.1-75.9 |
-
Note where (L & S gene): Cambodian = KC669690, KC669691& KC669694, KC669696; Thai = KC669692, KC669693 & KC669697, KC669698; Wenhzhou = KM386661, KM051421, KJ909795, KM051420 and KM386660, KM051423, KJ909794, and KM051422; Lassa = GU481076 & GU481077; Ippy = DQ328877 & DQ328878; Mopeia = DQ328874 & DQ328875; LCMV = AY847350 & AY847351; Junín = D10072 & AY216507; ORF = Open reading frame; NP = Nucleoprotein; GPC = Glycoprotein; nt = Nucleotide; aa = Amino acid; LCMV =: Lymphocytic Choriomeningitis Virus
Summary of PASC analysis.
| Sample ID | Country of origin | PASC: S segment | PASC: L segment | ||
|---|---|---|---|---|---|
| Sequence identity (%) | Closest virus | Sequence identity (%) | Closest virus | ||
| C0617 | Cambodia | 88.80 | Wēnzhōu virus | 86.27 | Wēnzhōu virus |
| C0649 | Cambodia | 88.51 | Wēnzhōu virus | 85.98 | Wēnzhōu virus |
| R4937 | Thailand | 70.63 | Wēnzhōu virus | 62.71 | Wēnzhōu virus |
| R5074 | Thailand | 70.29 | Wēnzhōu virus | 63.08 | Wēnzhōu virus |
-
Table 2—source data 1
PASC analysis.
- https://doi.org/10.7554/eLife.13135.005
Details of patient samples tested.
| Group | Clinical signs | Sample type (number of samples) | Collection dates | Collection locations | Mean age in years (range) | 95% CI (years) | Sex ratio | Test used | Number (proportion) of positive |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Dengue-like/ influenza-like illness | Acute and convalescent sera (n=510 including 98 acute, 214 convalescent and 198 paired sera) | 2005-2010 | Kampong Cham, various | 11.0 (4 months to 70 years) | 10.0-12.0 | 0.44 | IgG ELISA | 89 (17.4%) - Acute sera: 33 (33.7%) - Convalescence sera: 25 (11.7%) - Paired sera: 31 (15.7%) with evidence of seroconversion in 7 pairs (3.5%) |
| 2 | Healthy individuals (community dengue seroprevalence study) | Sera (n=529) | 2009 | Kampong Cham | 10.0 (1 month-20 years) | 9.59-10.42 | 0.50 | IgG ELISA | 70 (13.23%) |
| 3 | Meningo-encephalitis | Cerebrospinal fluid (n=200£) | 2013-2014 | Various | 6.5 (3 months-15 years) | 5.95-7.06 | 0.40 | Real Time RT-PCR | 0 (0%) |
| 4 | Dengue-like febrile illness | Sera collected during febrile stage (n=253*) | 2009, 2011-2013 | Various | 8.2 (1 to 38 years) | 7.6-8.8 | 0.48 | Semi-nested RT-PCR | 0 (0%) |
| 5a | Influenza-like illness (negative for four common respiratory viruses) | Nasopharyngeal swabs (n=328) | 2007-2012 | Various | 13.2 (1 month to 83 years) | 12.0-14.4 | 0.51 | Semi-nested RT-PCR | 4 (1.2%) |
| 5b | Influenza-like illness (positive for four common respiratory viruses) | Nasopharyngeal swabs (n=392#) | 0 (0%) | ||||||
| 6 | Acute lower respiratory infection | Nasopharyngeal swabs (n=279$) | 2008-2009 | Various | 2.7 (7 months to 63 years) | 2.1-3.4 | 0.41 | Semi-nested RT-PCR | 2 (0.7%) |
| 7 | Healthy individuals (H5N1 contacts) – negative control | Nasopharyngeal swabs (n=266§) | 2005-2011 | Various | 29.3 (1 to 79 years) | 27.1-31.5 | 0.45 | Semi-nested RT-PCR | 0 (0%) |
| 8 | Healthy anti-rabies vaccination volunteers – negative control | Nasopharyngeal swabs (n=238¥) | 2013 | Institute Pasteur Cambodia (Phnom Penh) | 26.7 (9 months to 83 years) | 24.5-28.8 | 0.56 | Semi-nested RT-PCR | 0 (0%) |
-
CI = Confidence Interval
-
£ 3 no information on age; * 8 no information on age and sex; # 2 no information on age and sex; $ 1 no information on age; § 5 no information on age and sex; 51 no information on sex; ¥ 5 no information on age and sex
Clinical details of human arenavirus infections.
| Case no. | Test used | Sex | Age | Hospitalised | Fever | Cough | Rhinorrhea | Nausea/ vomiting | Severe headache | Muscle pain | Other symptoms | Co-infection |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Screening nested RT-PCR | Female | 3 years | No | Yes | Yes | Yes | Human para-influenza virus 1 | ||||
| 2 | Screening nested RT-PCR | Male | 9 years | No | Yes | Yes | Yes | Yes | Yes | |||
| 3 | Screening nested RT-PCR | Male | 31 years | No | Yes | Yes | Yes | Yes | Yes | |||
| 4 | Screening nested RT-PCR | Female | 45 years | No | Yes | Yes | Yes | Yes | Yes | Yes | ||
| 5 | Screening nested RT-PCR | Female | 8 months | Yes | Yes | Yes | Dyspnea, wheezing, moderate anaemia (93g/L) | Unspecified Rhinovirus | ||||
| 6 | Screening nested RT-PCR | Male | 3 months | Yes | Yes | Yes | Dyspnea, wheezing, hypoglycaemia (28mmol/L) | Unspecified Rhinovirus | ||||
| S1 | ELISA | Male | 3 years | No | Yes | Yes | Yes | |||||
| S2 | ELISA | Male | 9 years | No | Yes | Yes | Yes | Yes | Yes | |||
| S3-7 | ELISA | No | Yes |
Additional files
-
Supplementary file 1
(A) Details of primers used for diagnostic RT-PCRs and genome sequencing. (B) Animals tested for arenavirus RNA by species and site, with number of positives shown in bold. (C) Details of rodent samples positive for arenavirus infection by screening RT-PCR .(D) Details of animals used for Cambodian virus infections. (E) Detailed IgG ELISA results for 7 patients with seroconversion. (F) Comparison between IFA and IgG ELISA results in human sera. (G) Arenavirus infections in patients with ILI symptoms who tested negative for 4 common respiratory viruses versus patients with ILI symptoms who tested positive and control group of healthy individuals. (H) Statistic analysis by age between respiratory illness group and healthy control group. (I) Multiple alignment of the sequences of the amplicons obtained by PCR for 3 patients. (J) Results of IgG ELISA in experimentally-infected rodents.
- https://doi.org/10.7554/eLife.13135.013





