Pre-transition effects mediate forces of assembly between transmembrane proteins
Figures
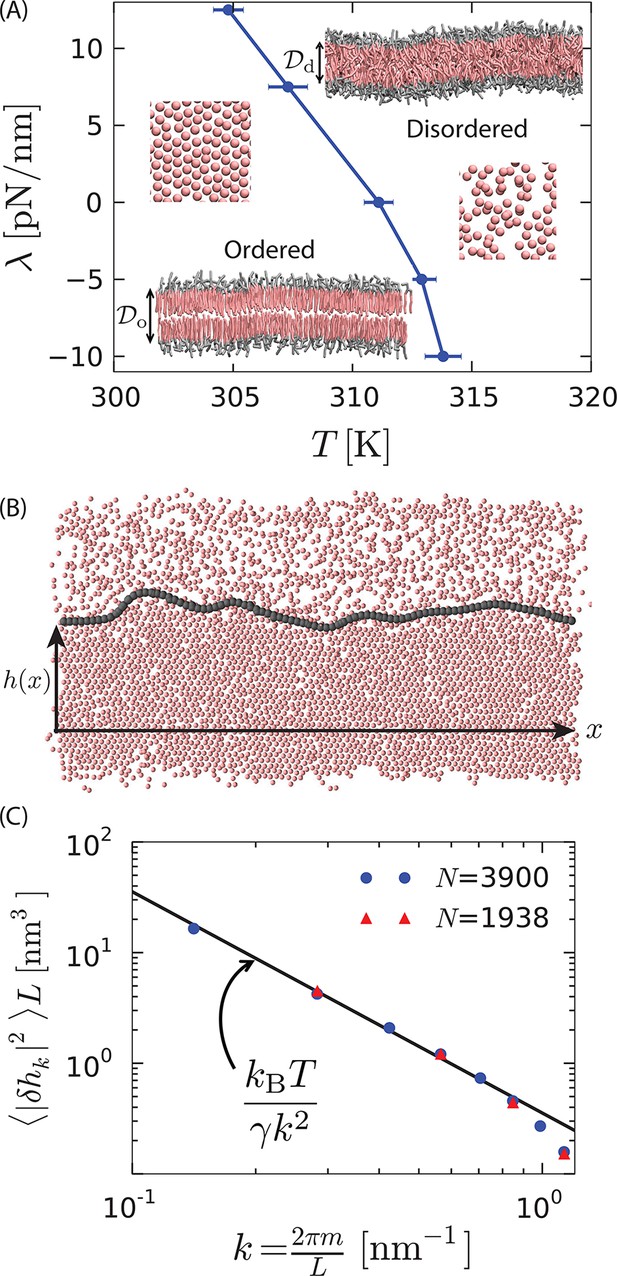
First-order phase transition in a model lipid bilayer.
(A) Order–disorder phase diagram in the tension–temperature, , plane. The lateral pressure across the membrane is . Points are estimated from 10 independent heating runs like those illustrated in Appendix 1–figure 1 for a periodic system with 128 lipids. Insets are cross sections showing configurations of a bilayer with 3200 lipids in the ordered and disordered phases. The heads are colored gray while the tails are colored pink. Water particles are omitted for clarity. The hydrophobic thicknesses, and , are the average vertical distances from the first tail particle of the upper monolayer to that of the lower monolayer. A macroscopic membrane buckles for all . Snapshots of the last tail beads in one monolayer of each phase are shown to illustrate the difference in packing. (B) Snapshot of a system showing coexistence between the ordered and disordered phases. The gray contour line indicates the location of the interface separating the ordered and disordered regions. The snapshot is a top view of the bilayer showing the tail-end particles of each lipid in one monolayer. is the distance of the instantaneous interface from a reference horizontal axis. (C) Fourier spectrum of . The line is the small- capillarity-theory behavior with pN.
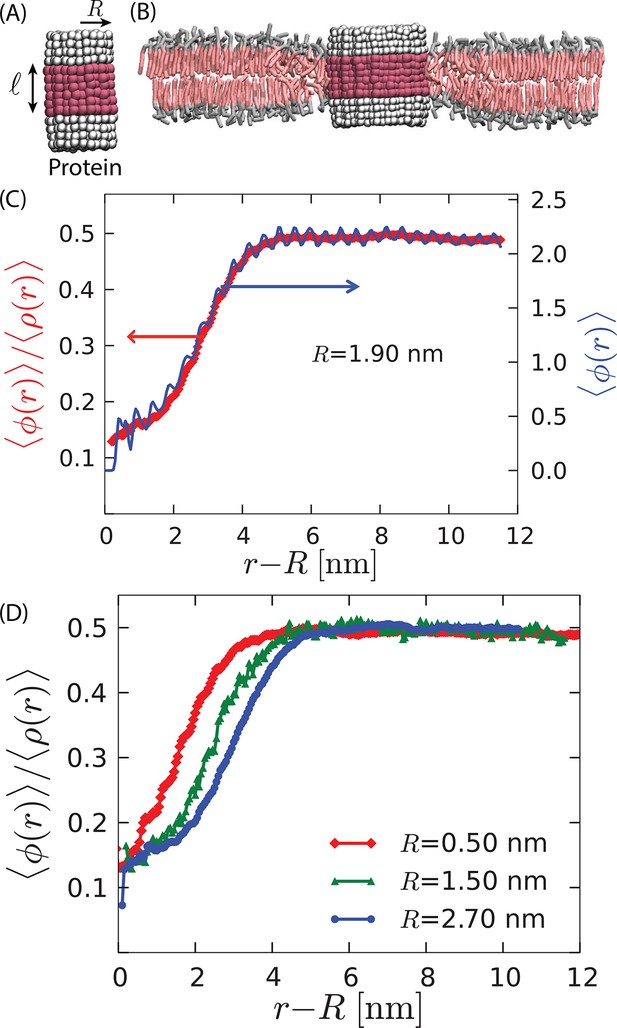
Model proteins in the bilayer.
(A) Idealized cylindrical protein-like solutes with radius and hydrophobic thickness (magenta). The hydrophilic caps of the protein are shown in white. (B) Cross section of the lipid bilayer in the ordered phase containing a model protein of radius 2.7 nm with a hydrophobic thickness nm . (C) The radial variation of the order parameters (right axis) and (left axis) show disorder in the vicinity of the protein of radius 1.9 nm. (D) Comparison of the radial order parameter variation for three different proteins shows an increase in the extent of the induced disorder region with protein radius.
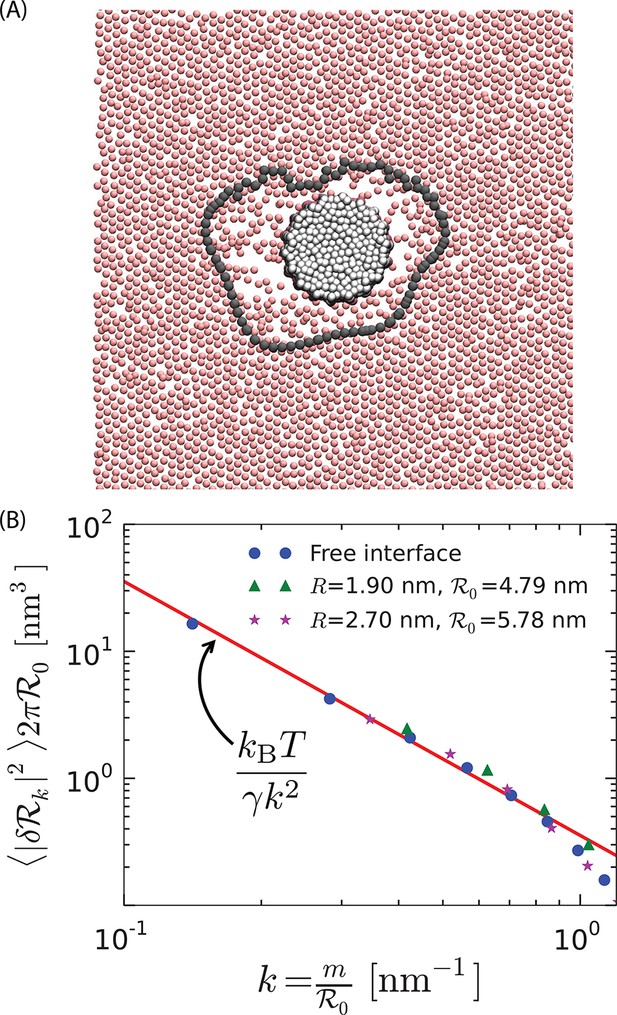
Soft order–disorder interface.
(A) Arrangement of the tail-end particles of the top monolayer corresponding to the protein in Figure 2B. Far away from the protein, the tail-end particles show hexagonal-like packing and are in the ordered state. Proximal to the protein, it can be seen that the tail-end particles are randomly arranged, and resemble the disordered phase. The line connected by the black points denotes the instantaneous order–disorder interface. (B) The fluctuations in the radius of the order–disorder interface are consistent with the fluctuations of a free order–disorder interface at coexistence. is the mean radius of the order–disorder interface surrounding a model protein of radius .
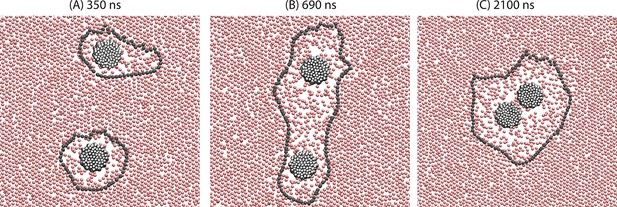
Demonstration of the orderphobic force: two proteins separated by a center-to-center distance of 14 nm are simulated at 309 K.
Snapshots at various times reveal the process of assembly in which the two order–disorder interfaces merge into a single interface.
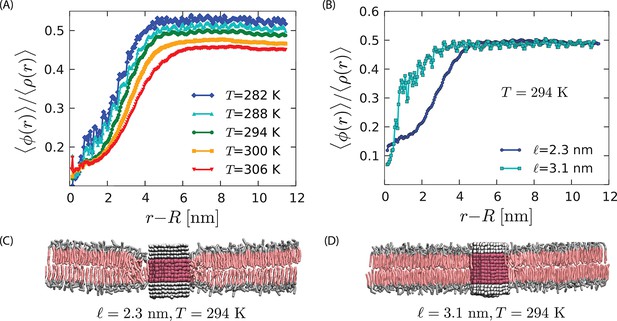
Strength of the orderphobic force.
(A) Radial variation of the order parameter showing the extent of the disordered region as a function of temperature, for a protein of radius 1.9 nm and hydrophobic thickness 2.3 nm. The extent of the disordered region increases as the melting temperature is approached, at zero surface tension. (B) Comparison of the radial variation of the order parameter for different hydrophobic mismatches. Proteins with no mismatch do not create any disordered region. (C) Arrangement of lipids around a protein with negative mismatch. (D) Arrangement of lipids around a protein with zero mismatch.
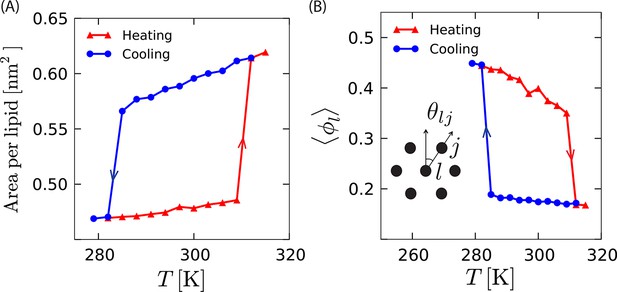
Structural measures of different phases as a function of temperature, .
(A) Variation in area per lipid with temperature during heating and cooling shows finite jumps and hysteresis. (B) Average local orientational order, , also shows finite jumps as a function of temperature while heating and cooling. Magnitudes of heating and cooling rates are 3 K/µs.
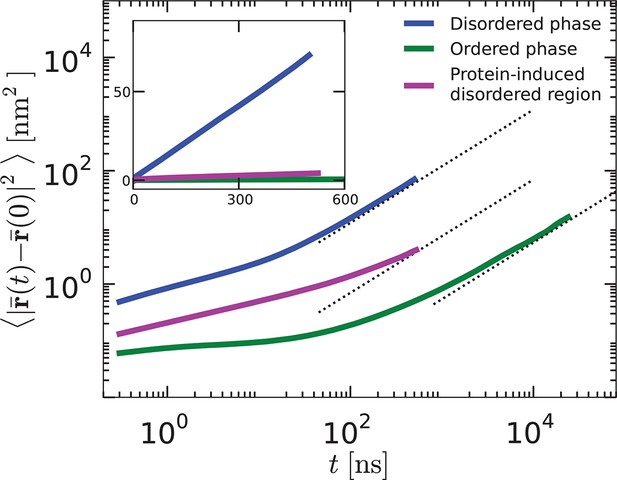
Mean-square displacements as functions of time, , for lipids in the disordered phase, the protein-induced disordered domain, and the ordered phase.
The functions are shown on log–log scale (main graphs) and linear scales (inset).
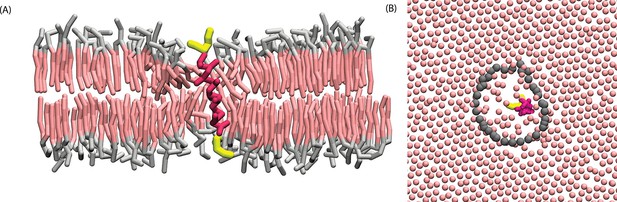
A model -helix is orderphobic.
(A) Cross section of ordered phase of a hydrated DPPC membrane containing one transmembrane KALP23 protein. Solvent water is not rendered for purpose of clarity. Configuration was obtained after running simulation for roughly 1 µs at 294 K and zero lateral pressure. (B) Configuration of tail-end particles for the top monolayer, with gray points locating the instantaneous interface.
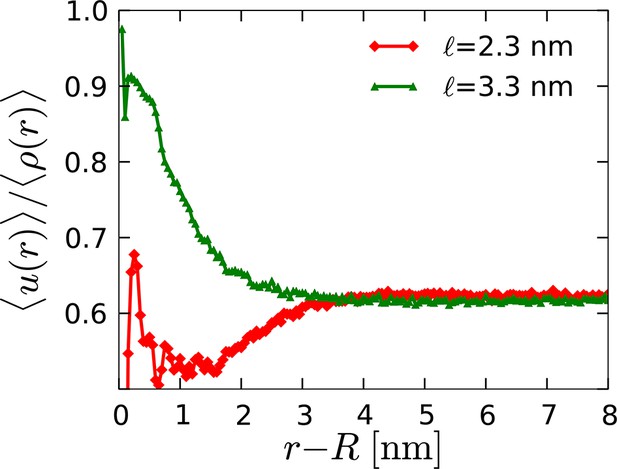
Radial profile of the average director density surrounding orderphobic ( nm) and orderphilic ( nm) model proteins in the disordered membrane phase.
https://doi.org/10.7554/eLife.13150.013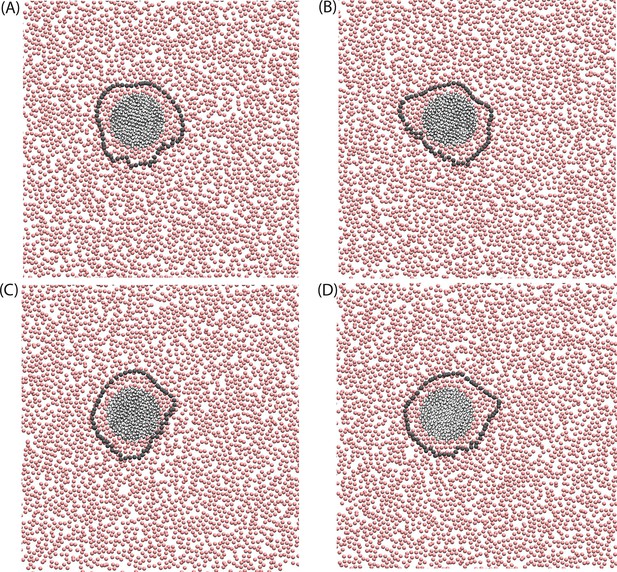
Configurations of the disordered membrane in the presence of a model orderphilic protein, nm.
The rendered particles are the ‘C2’ tail particles of the lipids, and the gray line marks the boundary between ordered and disordered domains by rendering the contour of the instantaneous interface for the director field, .
Videos
Instantaneous interface around an orderphobic protein.
Also uploaded to https://goo.gl/NBQJP9.
Assembly of two orderphobic proteins.
Also uploaded to https://goo.gl/HXS0j7.





