Comparative genomics explains the evolutionary success of reef-forming corals
Figures
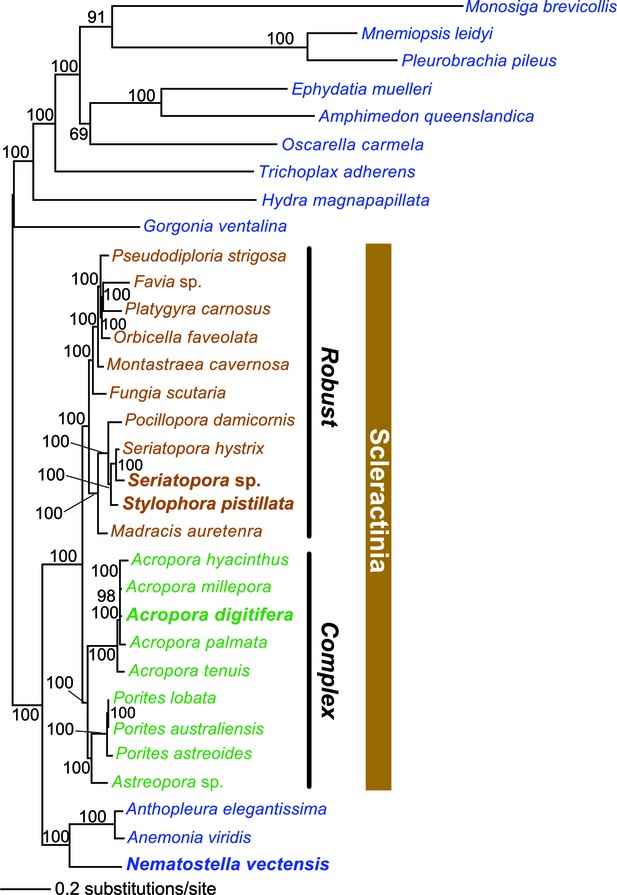
Multigene maximum likelihood (RAxML) tree inferred from an alignment of 391 orthologs (63,901 aligned amino acid positions) distributed among complete genome (boldface taxon names) and genomic data from 20 coral species and 12 outgroups.
The PROTGAMMALGF evolutionary model was used to infer the tree with branch support estimated with 100 bootstrap replicates. Robust and complex corals are shown in brown and green text, respectively, and non-coral metazoan species are shown in blue text.
-
Figure 1—source data 1
Coral genomic data compiled in this study and their attributes.
- https://doi.org/10.7554/eLife.13288.004
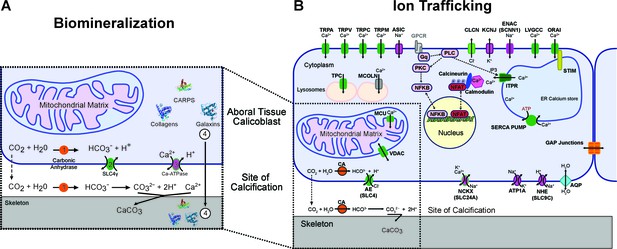
The mechanism of (A) coral biomineralization based on data from physiological and molecular approaches and (B) the major components of the human ion trafficking system that were identified in the coral genomic data (Figure 2—source data 1 for details).
Here, in (A) Biomineralization, 1 = carbonic anhydrases (orange); 2 = bicarbonate transporter (green); 3 = calcium-ATPase (purple); 4 = organic matrix proteins (shown as protein structures).
-
Figure 2—source data 1
Major components of the human ion trafficking system identified in the coral genomic data.
- https://doi.org/10.7554/eLife.13288.006
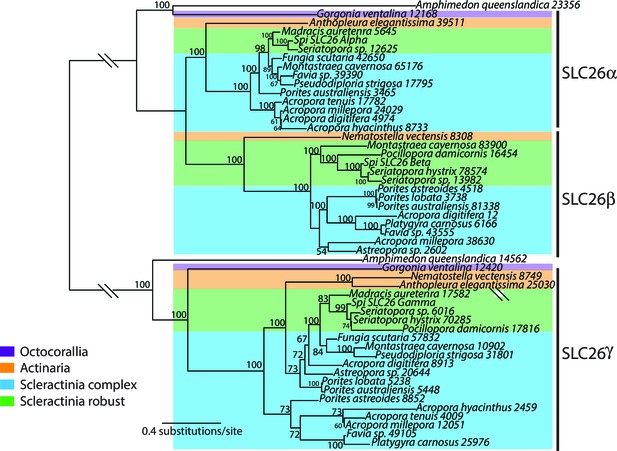
Bayesian consensus trees of SLC26.
Bayesian posterior probabilities are indicated when greater than 50%. For this analysis and for the trees shown in Figure 2—figure supplements 2–4, MrBayes v3.1.2 was used with a random starting tree and the LG model of amino acid substitution. Trees were generated for 6,000,000 generations and sampled every 1000 generations with four chains to obtain the consensus tree and to determine the posterior probabilities at the internal nodes.
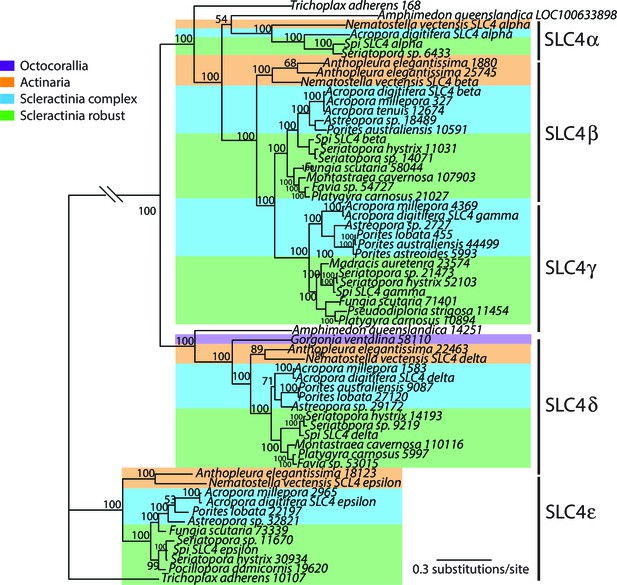
Bayesian consensus trees of SLC4.
Bayesian posterior probabilities (×100) are indicated when greater than 50%.
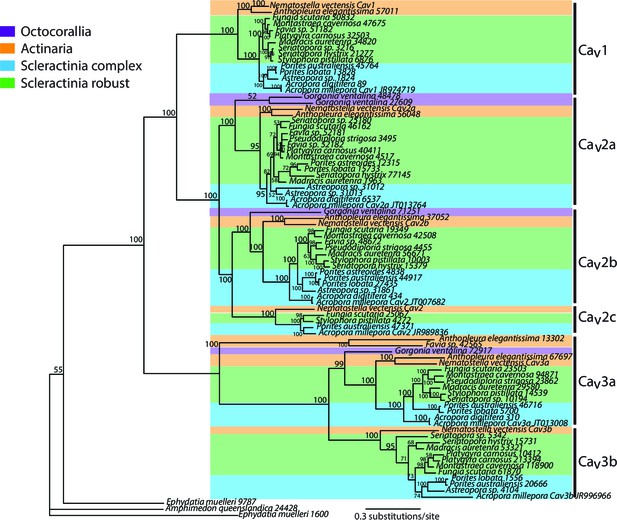
Bayesian consensus trees of Cav.
Bayesian posterior probabilities (×100) are indicated when greater than 50%.
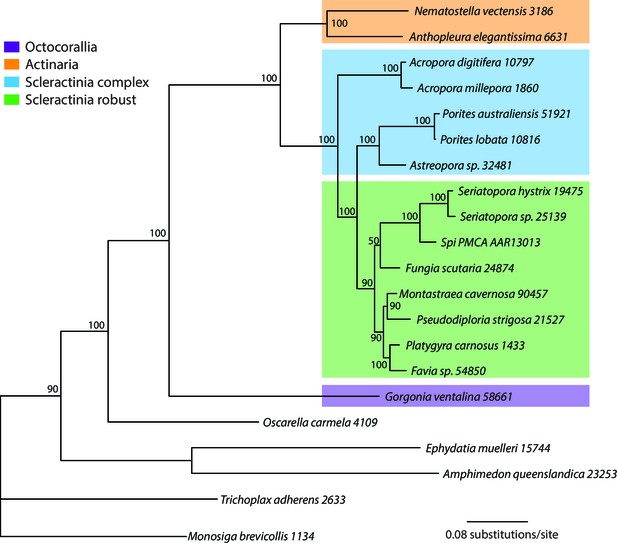
Bayesian consensus trees of coral and outgroup Ca-ATPase proteins.
Bayesian posterior probabilities (×100) are indicated when greater than 50%.
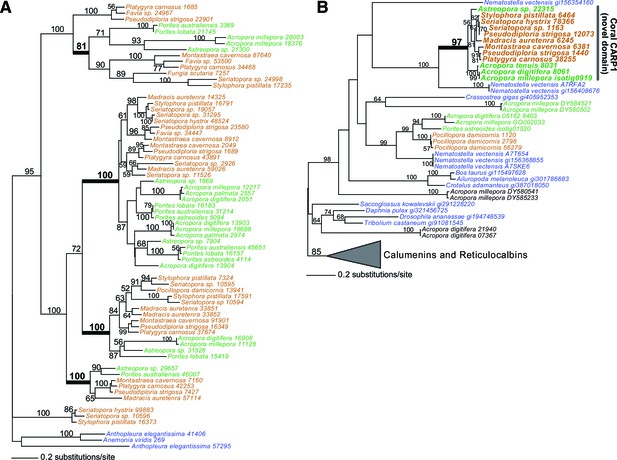
Evolution of CARPs and other coral acid-rich proteins.
(A) Maximum likelihood (RAxML) tree showing extensive history of duplication of genes encoding CARP 5 that predates the split of robust (brown text) and complex (green text) corals. (B) RAxML tree showing the origin of CARP 1 in robust (brown text) and complex (green text) corals from a reticulocalbin-like ancestor by the evolution of a novel acid-rich N-terminaldomain. The non-coral species in both trees are shown in blue text.
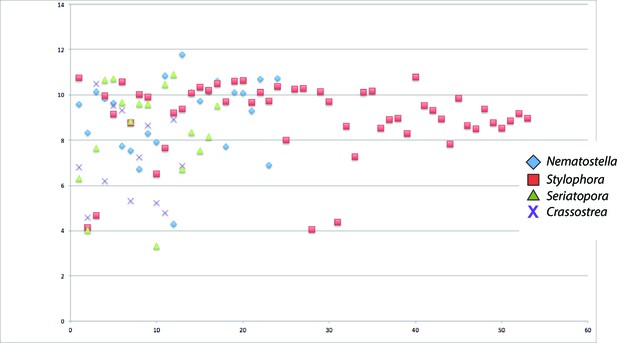
Scatter plot of isoelectric points of collagens from Seriatopora, Stylophora, Nematostella, and Crassostrea gigas.
https://doi.org/10.7554/eLife.13288.012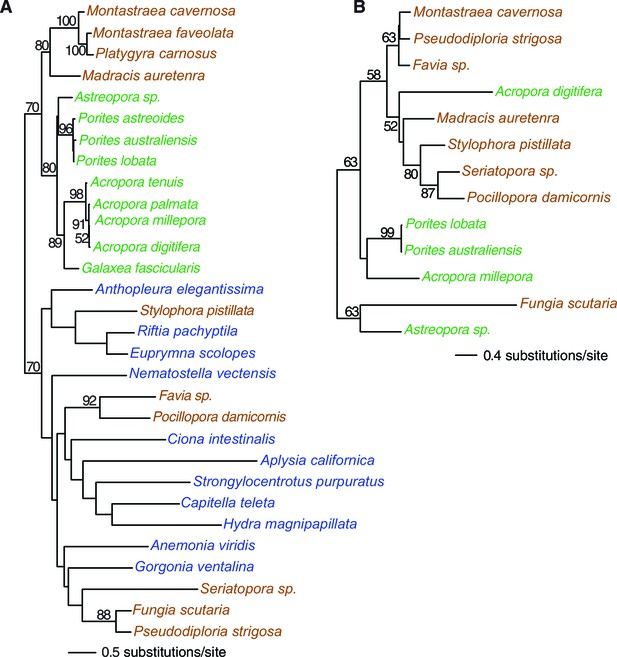
Maximum likelihood (ML) trees of galaxin and amgalaxin.
(A) ML tree of best galaxin hits from 19 coral species (brown for robust corals and green for complex corals) and 11 non-coral species (blue text). (B) ML tree of best amgalaxin hits from 13 coral species. No outgroup blast hits were found against the acidic region of Acropora millepora amgalaxin 1 or 2 (Genbank accession numbers ADI50284.1 and ADI50285.1, respectively).
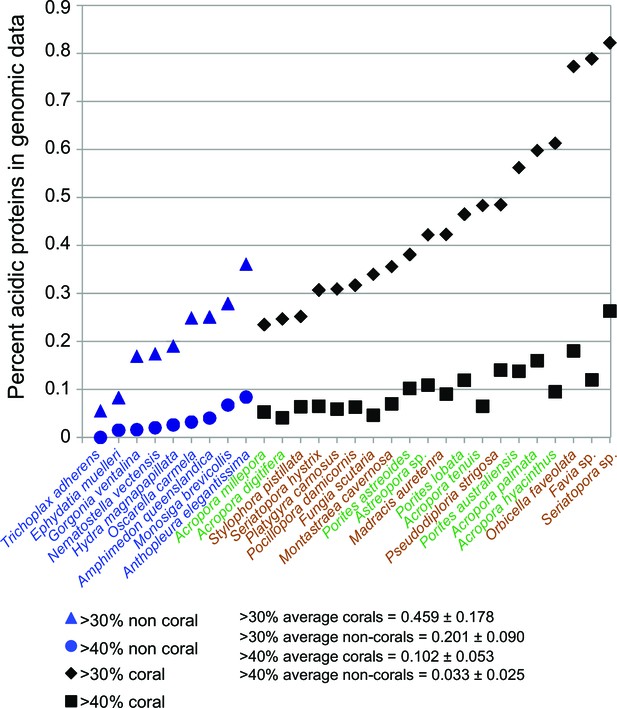
Comparison of robust coral (brown text) and complex coral (green text) and non-coral (blue text) genomes with respect to percent of encoded proteins that contain either >30% or >40% negatively charged amino acid residues (i.e., aspartic acid [D] and glutamic acid [E]).
The average composition and standard deviation of D + E is shown for the two cut-offs of these estimates. On average, corals contain >2-fold more acidic residues than non-corals. This acidification of the coral proteome is postulated to result from the origin of biomineralization in this lineage.

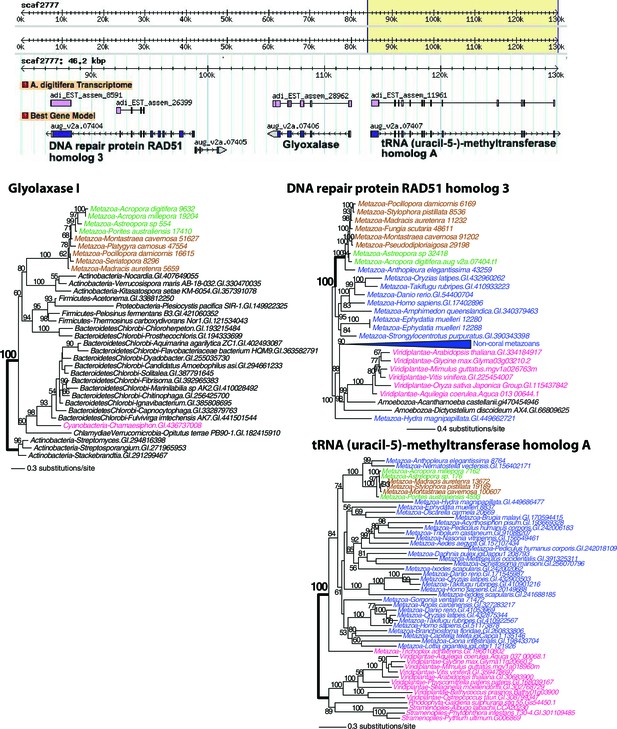
Analysis of a genomic region in Acropora digitifera that encodes a putative HGT candidate.
(A) The genome region showing the position of the HGT candidate (PNK3P) and its flanking genes. (B) Maximum likelihood trees of PNK3P (polynucleotide kinase 3 phosphatase, pfam08645) domain-containing protein and the proteins (RNA-binding and GTP-binding proteins) encoded by the flanking genes. Robust and complex corals are shown in brown and green text, respectively, and non-coral metazoan and choanoflagellate species are shown in blue text. Photosynthetic lineages, regardless of phylogenetic origin, are shown in magenta text and all other taxa are in black text. GenBank accession (GI) or other identifying numbers are shown for each sequence. The PNK3P domain plays a role in the repair of DNA single-strand breaks by removing single-strand 3'-end-blocking phosphates (Petrucco et al., 2002).

Maximum likelihood trees of a DEAD-like helicase and the protein encoded by the flanking gene.
The bacterium-derived DEAD-like helicase genes in coral are nested within bacterial sequences, whereas the upstream host-derived gene (encoding mannosyl-oligosaccharide 1,2-alpha-mannosidase IB) is monophyletic with homologous genes from other metazoan species. The downstream Acropora digitifera-specific gene has no detectable homolog in other species. Robust and complex corals are shown in brown and green text, respectively, and non-coral metazoan and choanoflagellate species are shown in blue text. Photosynthetic lineages, regardless of phylogenetic origin, are shown in magenta text and all other taxa are in black text. GenBank accession (GI) or other identifying numbers are shown for each sequence.
Maximum likelihood tree of an exonuclease-endonucease-phosphatase (EEP) domain-containing protein (A), an ATP-dependent endonuclease (B), a tyrosyl-DNA phosphodiesterase 2-like protein (C), and DNA mismatch repair (MutS-like) protein (D).
Robust and complex corals are shown in brown and green text, respectively, and non-coral metazoan and choanoflagellate species are shown in blue text. Photosynthetic lineages, regardless of phylogenetic origin, are shown in magenta text and all other taxa are in black text. GenBank accession (GI) or other identifying numbers are shown for each sequence.

Maximum likelihood trees of glyoxalase I (or lactoylglutathione lyase) and the proteins encoded by the flanking genes (top image) in Acropora digitifera.
Robust and complex corals are shown in brown and green text, respectively, and non-coral metazoan and choanoflagellate species are shown in blue text. Photosynthetic lineages, regardless of phylogenetic origin, are shown in magenta text and all other taxa are in black text. GenBank accession (GI) or other identifying numbers are shown for each sequence.
Maximum likelihood tree of a second glyoxalase I (or lactoylglutathione lyase) and the proteins encoded by the flanking genes (top image) in Acropora digitifera.
The coral glyoxalase gene gene was derived from a bacteria-specific gene type. Robust and complex corals are shown in brown and green text, respectively, and non-coral metazoan and choanoflagellate species are shown in blue text. Photosynthetic lineages, regardless of phylogenetic origin, are shown in magenta text and all other taxa are in black text. GenBank accession (GI) or other identifying numbers are shown for each sequence.
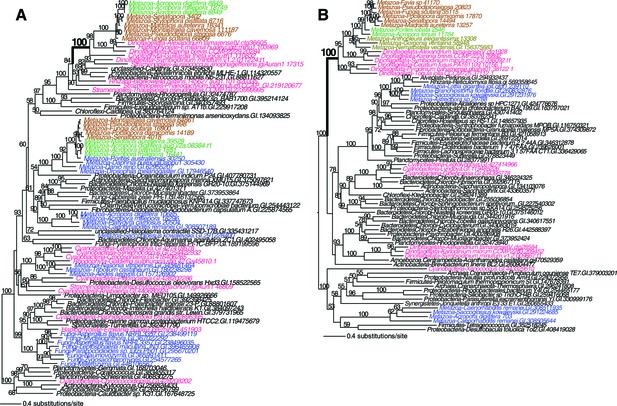
Maximum likelihood tree of an algal-derived short-chain dehydrogenase/reductase (A), and a dinoflagellate-derived phosphonoacetaldehyde hydrolase (B).
Robust and complex corals are shown in brown and green text, respectively, and non-coral metazoan and choanoflagellate species are shown in blue text. Photosynthetic lineages, regardless of phylogenetic origin, are shown in magenta text and all other taxa are in black text. GenBank accession (GI) or other identifying numbers are shown for each sequence.
Tables
The list of non-redundant anthozoan genes derived via HGT.
| No. | Ancestor | Genes | Protein products | Support | Source(s) |
|---|---|---|---|---|---|
| 1 | Coral | A. digitifera_2036 | PNK3P | 100 | CA |
| 2 | Coral | A. digitifera_8849 | SDR | 100 | CA |
| 3 | Coral | Seriatopora_31861 | DEAD-like helicase | 100 | Bact |
| 4 | Coral | Seriatopora_16594 | glyoxalase | 100 | CA |
| 5 | Coral | Seriatopora_17147 | acyl- dehydrogenase | 100 | Bact |
| 6 | Coral | Seriatopora_17703 | carbonic anhydrase | 96 | Dino |
| 7 | Coral | Seriatopora_19477 | fatty acid or sphingolipid desaturase | 100 | CA |
| 8 | Coral | Seriatopora_3957 | atpase domain-containing protein | 100 | Bact |
| 9 | Coral | Seriatopora_7060 | sam domain-containing protein | 100 | Bact |
| 10 | Coral | Seriatopora_7928 | atp phosphoribosyltransferase | 100 | CA/Fungi |
| 11 | Coral | Seriatopora_8296 | glyoxalase | 98 | Bact |
| 12 | Coral | Seriatopora_22596 | 2-alkenal reductase | 92 | Bact |
| 13 | Coral | Seriatopora_28321 | histidinol-phosphate aminotransferase | 96 | Unclear |
| 14 | Anthozoa | A. digitifera_418 | duf718 domain protein | 100 | CA |
| 15 | Anthozoa | A. digitifera_15871 | peptidase s49 | 96 | Algae/Bact |
| 16 | Anthozoa | A. digitifera_14520 | predicted protein | 100 | CA/Bact |
| 17 | Anthozoa | A. digitifera_7178 | rok family protein/fructokinase | 93 | Red algae |
| 18 | Anthozoa | A. digitifera_10592 | Phospholipid methyltransferase | 100 | CA/Viri |
| 19 | Anthozoa | A. digitifera_13390 | predicted protein | 100 | Bact |
| 20 | Anthozoa | A. digitifera_313 | malate synthase | 98 | CA/Bact |
| 21 | Anthozoa | A. digitifera_1537 | hypothetical protein | 100 | Bact |
| 22 | Anthozoa | A. digitifera_13577 | gamma-glutamyltranspeptidase 1-like | 100 | Unclear |
| 23 | Anthozoa | A. digitifera_5099 | Isocitrate lyase (ICL) | 100 | Bact |
| 24 | Anthozoa | A. digitifera_13467 | uncharacterized iron-regulated protein | 100 | CA |
| 25 | Anthozoa | A. digitifera_6866 | 3-dehydroquinate synthase | 98 | CA |
| 26 | Anthozoa | A. digitifera_11675 | intein c-terminal splicing region protein | 100 | Bact |
| 27 | Anthozoa | Seriatopora_10994 | penicillin amidase | 100 | Bact |
| 28 | Anthozoa | Seriatopora_14009 | nucleoside phosphorylase-like protein | 100 | Bact |
| 29 | Anthozoa | Seriatopora_14494 | phosphonoacetaldehyde hydrolase | 100 | Dino |
| 30 | Anthozoa | Seriatopora_15303 | exonuclease-endonuclease-phosphatase | 99 | CA/Viri |
| 31 | Anthozoa | Seriatopora_15772 | fmn-dependent nadh-azoreductase | 99 | Dino |
| 32 | Anthozoa | Seriatopora_19888 | had family hydrolase | 97 | Algae/Bact |
| 33 | Anthozoa | Seriatopora_20039 | chitodextrinase domain protein | 92 | Dino |
| 34 | Anthozoa | Seriatopora_20146 | glutamate dehydrogenase | 100 | CA/Bact |
| 35 | Anthozoa | Seriatopora_20479 | thif family protein | 100 | Bact |
| 36 | Anthozoa | Seriatopora_21195 | ATP-dependent endonuclease | 100 | Dino |
| 37 | Anthozoa | Seriatopora_8585 | chitodextrinase domain protein | 92 | Bact |
| 38 | Anthozoa | Seriatopora_24047 | aminotransferase | 100 | Bact |
| 39 | Anthozoa | Seriatopora_25961 | d-alanine ligase | 99 | Bact |
| 40 | Anthozoa | Seriatopora_26478 | quercetin 3-o-methyltransferase | 100 | Viri |
| 41 | Anthozoa | Seriatopora_29443 | diaminopimelate decarboxylase | 100 | CA |
-
Bact: Bacteria; CA: chlorophyll c-containing algae; Dino: dinoflagellates; Viri: Viridiplantae.
Additional files
-
Supplementary file 1
Taxonomic compilation and presence/absence in each taxon for genes involved in oxidative stress, DNA repair, cell cycle and apoptosis.
The values in parentheses show the number of taxa in which the gene sequence was recovered in the genomic database.
- https://doi.org/10.7554/eLife.13288.022





