Structural basis for DNA 5´-end resection by RecJ
Figures
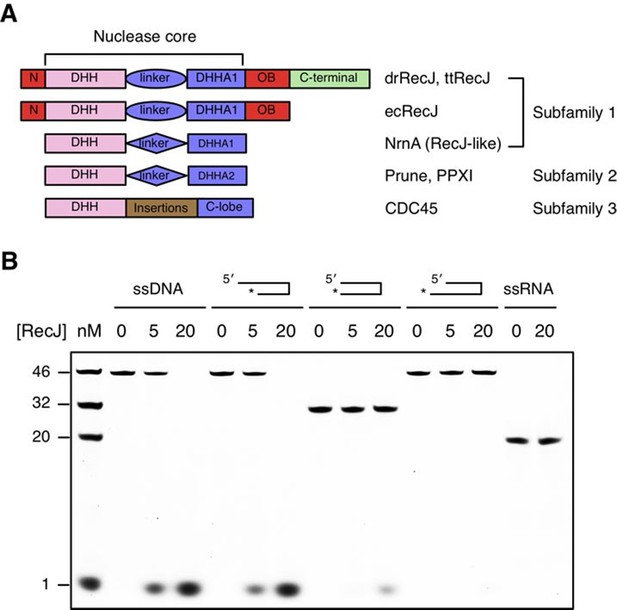
Domain arrangement and substrate specificity of drRecJ.
(A) Schematic of the domain arrangements of three DHH subfamilies. (B) Denaturing PAGE gel showing that drRecJ degrades different substrates, as shown at the top of the panel. 3′-Fluorescent labeled DNA or RNA (100 nM) were incubated with drRecJ (0, 5 and 20 nM) in the presence of 100 nM Mn2+ (see methods).
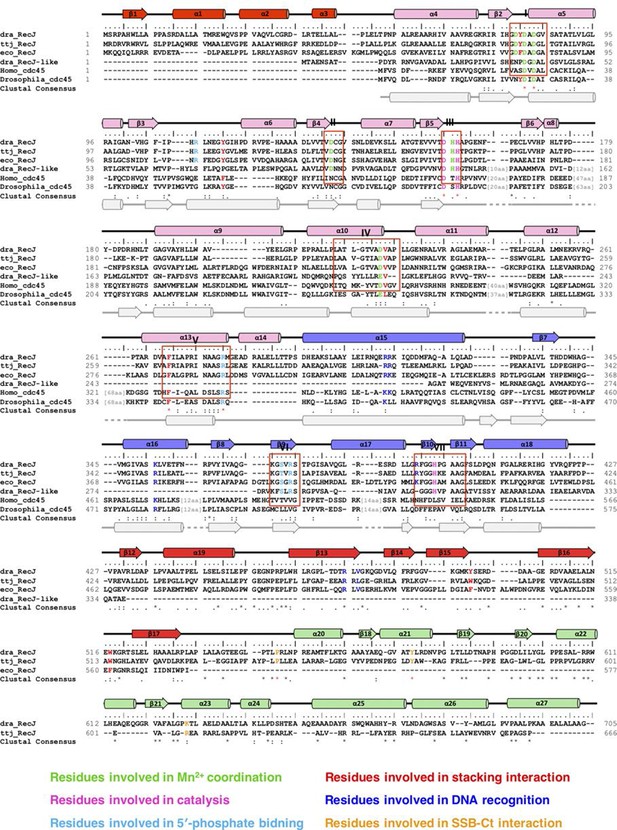
Sequence alignments, secondary structure, and functional residues of RecJ and CDC45.
Names of species are dra, Deinococcus radiodurans; ttj, Thermus thermophilus; eco, Escherichia Coli; Homo, Homo sapiens; Drosophila, Drosophila melanogaster. Structural elements of RecJ are shown in distinct colors. Predicted secondary structure of human Cdc45 is shown at bottom (grey). Conserved motifs (I-VII) are labeled. Conserved key residues are highlighted in distinct colors.
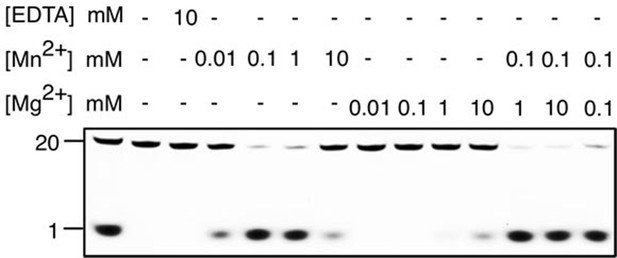
Metal preference of drRecJ.
3′-fluorescent labeled 20 nt ssDNA (KY08, 100 nM) was incubated with drRecJ (10 nM) in the presence of Mn2+ and Mg2+. For the metal competition assays, drRecJ was pre-incubated with 0.1 mM Mn2+ before addition of Mg2+.
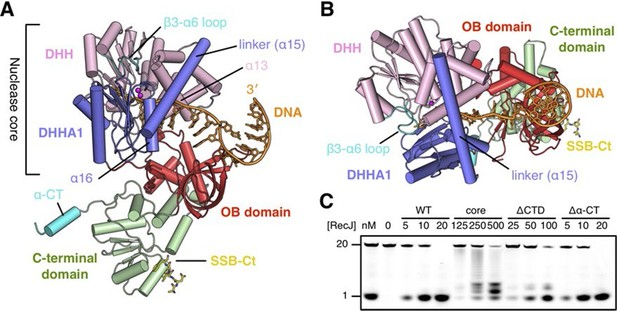
Structure of drRecJ complex.
(A) Overall structure of drRecJ complex viewed from the side. Protein domains of drRecJ are labeled and shown in distinct colors. The DNA and SSB-Ct are colored orange and yellow, respectively. Two Mn2+ in the active site are shown as magenta spheres. Two regions that are disordered in the ttRecJ structures (PDB code: 2ZXP) are highlighted in cyan. Three helices that form a helical gateway are also labeled. (B) Overall structure of the drRecJ complex viewed from the top of the DNA. The downstream nucleotides stack well to mimic the double-stranded DNA. (C) Denaturing PAGE gel showing the nuclease activities of different truncations of drRecJ. 3′-Fluorescence-labeled 20 nt ssDNA (100 nM) was incubated with various concentrations of different truncations of drRecJ proteins (see methods).
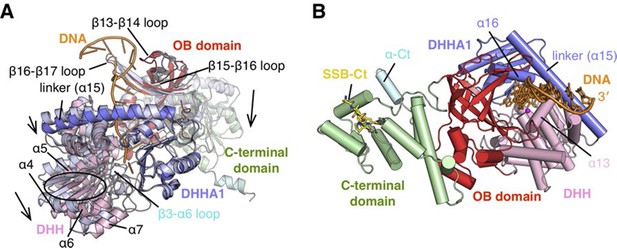
Structure of RecJ:DNA complex.
(A) Comparison of complex II and ttRecJ structures. The ttRecJ (PDB code: 2ZXP) is colored white. Protein domains of drRecJ are shown in distinct colors and labeled. The DNA and ordered β3-α6 loop are colored orange and cyan, respectively. Loops and helices showing noticeable deviations are labeled. The relative domain movements are shown by the arrowheads. (B) Overall structure of complex II viewed from the top of the DNA entrance. Protein domains of drRecJ are shown in distinct colors and labeled. The DNA and SSB-Ct are colored orange and yellow, respectively. Two Mn2+ in the active site are shown as magenta spheres. Three helices (α13, α15 and α16) that form a helical gateway are also labeled.
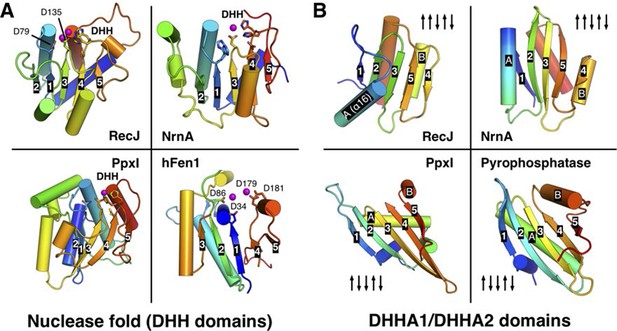
Comparison of the nuclease fold and DHHA1/DHHA2 domain.
(A) Structural comparison among the nuclease fold from drRecJ, NrnA (PDB code: 4LS9), PpxI (PDB code: 2QB6) and hFen1 (PDB code: 3Q8K). The nuclease fold of each enzyme is shown in rainbow colors from the blue N- to the red C-terminus. The residues involved in catalysis are shown as sticks. The topologies of β-strands are also labeled. (B) Structural comparison between the DHHA1 domain (drRecJ, NrnA) and the DHHA2 domain (PpxI, Pyrophosphatase (PDB code: 1WPM)). The topologies of β-strands and two inserted α-helices (A and B) are labeled.
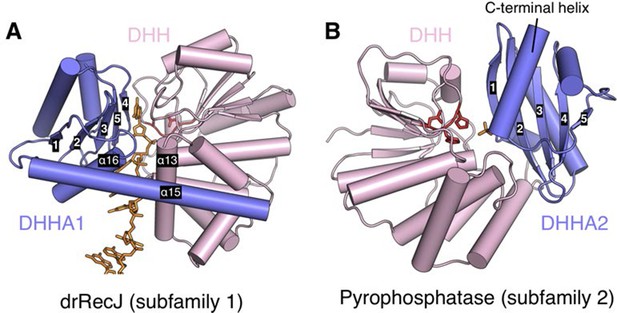
Relative position of DHH (pink) and DHHA1/DHHA2 (blue) domain.
The active site of subfamily 1 group is formed between DHH domain and β3 to β5 of DHHA1 domain (A). While the β1 of DHHA2 domain and DHH domain form the active site of subfamily 2 group proteins (B). DNA and phosphate group are colored orange and DHH motif is shown as red stick. The helical gateway in DHHA1 domain and C-terminal α-helix in DHHA2 are also labeled.
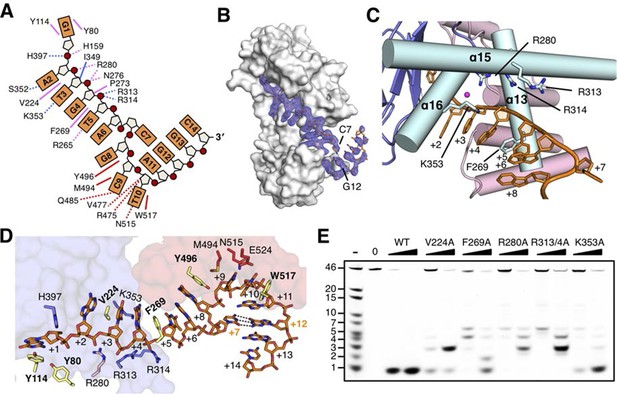
The DNA binding in the RecJ-DNA complex.
(A) Schematic of the numbered DNA substrate used for complex II crystallization. Interactions between nucleotides and DHH domain, DHHA1 domain and OB fold domain are colored pink, blue and red, respectively. Hydrogen bonds are defined as within 3.2 Å and van der Waals contacts within 4.2 Å (dashed lines). Solid lines indicate residues that stack with DNA bases. (B) drRecJ surface and 2Fo−Fc electron density of DNA contoured at 1σ. The C7-G12 base pair is labeled. (C) The helical gateway is labeled and shown in cyan. Key residues interacting with DNA are labeled and shown as sticks. Nucleotides are labeled as in (A). (D) Interactions between drRecJ and DNA in complex II structure. Protein side chains involved in the protein-DNA interactions are shown as sticks, and the key residues that form stacking interactions are highlighted in yellow. Nucleotides are labeled as in (A). C7 and G12 (orange) form Watson-Crick base pair, as indicated by the dark dashed line. (E) Denaturing PAGE gel showing the reduced nuclease activity and processivity of mutant drRecJ proteins (alanine substitutions of key residues involved in DNA binding). 3′-Fluorescence-labeled 46 nt ssDNA (100 nM) was incubated with drRecJ proteins (0, 5 and 20 nM) in the presence of 100 nM Mn2+ (see methods).
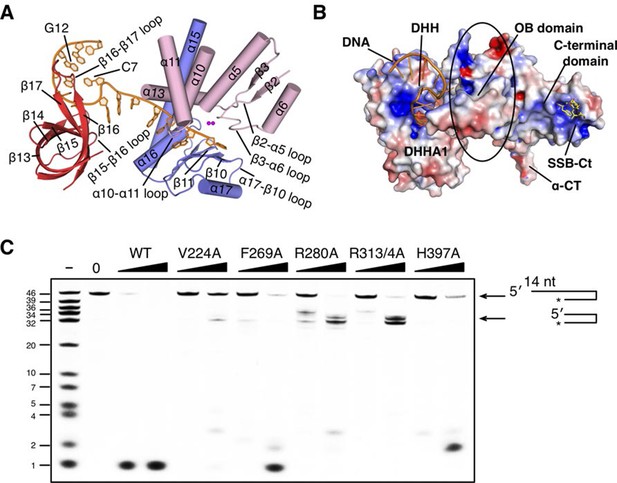
The DNA binding of drRecJ.
(A) Key structural elements in complex II. Structural elements of DNA binding and metal ions are indicated and colored according to the scheme in Figure 2A. Loops involved in nucleotide binding are also indicated. (B) The distribution of the electrostatic surface of drRecJ. Blue and red represent the positive and negative charge potential at the + and −10 kTe−1 scale, respectively. Electropositive patch were observed as DNA and SSB-Ct binding sites. (C) Denaturing PAGE gel showing that mutations of key residues involved in nuclease core-DNA interactions impaired the nuclease activity, processivity and the digestion of DNA with 5´-ssDNA overhang (stops at ss-dsDNA junction). For the reaction, 3′-fluorescent labeled DNA containing 5′-ssDNA overhang (KY04, 100 nM) were incubated with wild-type or mutant drRecJ proteins (10 and 20 nM) in the presence of 100 nM Mn2+.
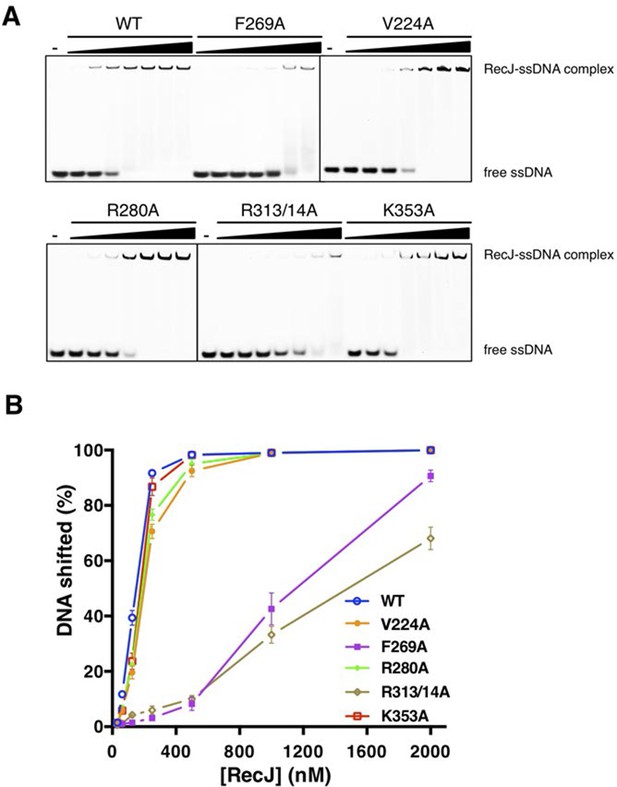
DNA binding activity of drRecJ mutant proteins as in the Figure 3E.
(A) Electrophoretic mobility shift assays were performed with 100 nM 3′-fluorescent labeled 20 nt poly (dA) and different concentrations of RecJ (31.5, 62.5, 125, 250, 500, 1000 and 2000 nM). (B) A plot of quantified relative band intensities of the RecJ-DNA complex bands from (A). Data are represented as mean ± SEM.
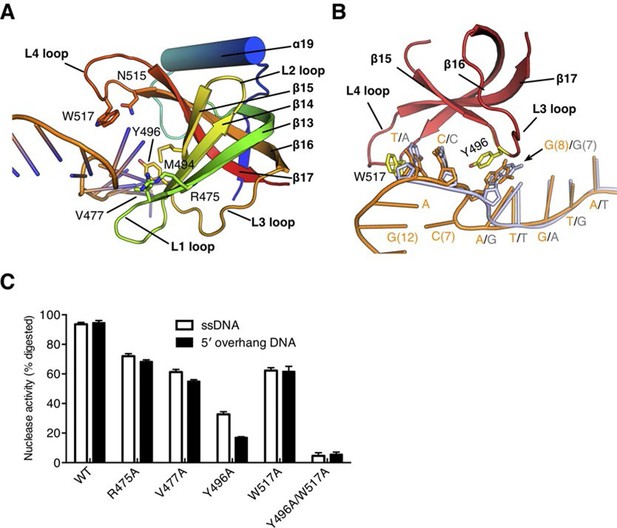
The OB fold domain is critical for drRecJ resection.
(A) The OB fold domain is shown in rainbow-colored diagrams. Residues involved in DNA binding are labeled and shown as sticks. (B) A comparison of DNA in the complex II (DNA with 5´-ssDNA overhang; orange) with complex III (ssDNA; white). Two conserved aromatic residues Tyr496 and Trp517 are shown as sticks (yellow). The black arrowhead indicates the position of stacking interaction between Tyr496 and the guanine base. (C) Quantification and plot of ssDNA and DNA with 5´-ssDNA overhang, which are processed by wild-type and mutant drRecJ proteins (alanine substitutions of key residues in the OB fold domain), using the same DNA substrate and reaction conditions as in Figure 1B. Data are represented as mean ± SEM.
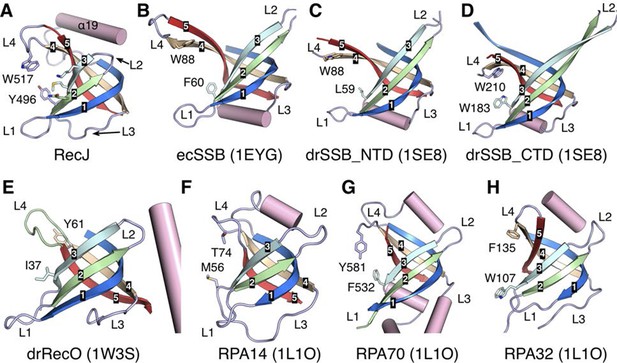
Structural comparison of the OB fold from E. coli SSB (ecSSB), D. radiodurans SSB subunits (drSSB_NTD and drSSB_CTD), D. radiodurans RecO (drRecO) and human replication protein A subunits (RPA14, RPA32 and RPA70).
Their PDB accession codes are shown in parentheses and references can be found in the main text. The mixed five β-strands are labeled and shown in distinct colors. Four loops are labeled as L1-L4. Conserved aromatic residues in loop3 and loop4 are shown as sticks.
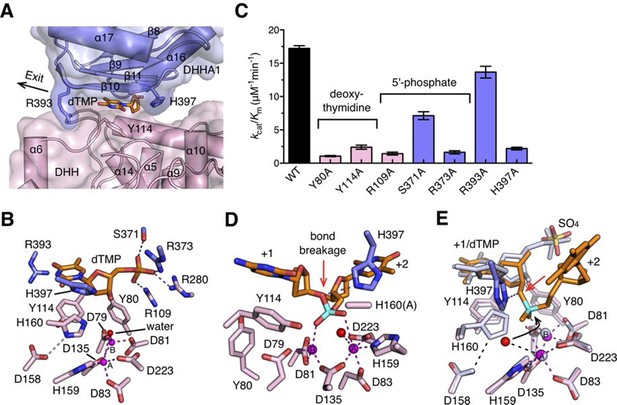
Nucleotide binding site and the catalysis.
(A) dTMP binding pocket between the DHH domain and DHHA1 domain. dTMP is shown as stick (orange). Two conserved residues His397 and Arg393 at the entrance and exit to the active site are labeled and shown as sticks. (B) Close-up of the active site of drRecJ-dTMP (complex I). Two Mn2+ ions (A and B; magenta) are coordinated by a water molecule (red sphere) and conserved Asp and His residues (magenta dashed lines). Asp158 forms a hydrogen bond with His160, as indicated by the pink dashed line. The phosphate of the dTMP is held by Arg109, Arg280, Ser371 and Arg373 (blue dashed lines). (C) Catalytic efficiency of wild-type and mutant drRecJ as a bar graph showing the relative severity of the mutations. The Michaelis-Menten kinetics data are from Table 2. Data are represented as mean ± SEM. (D) Close-up of the active of site of complex III. The scissile phosphate centered on two catalytic Mn2+ is highlighted in cyan. The red arrowhead indicates the position of P-O bond breakage. Interaction between His397 and the oxygen atom of the scissile phosphate group is indicated by blue dashed line. (E) A comparison of the active sites in the complex I (white) with complex III (same color as in panel D). The nucleophilic water molecule observed in complex III occupies the position close to the His160 in drRecJ-dTMP structure (complex I). The red arrowhead indicates the position of P-O bond breakage and the black arrowhead indicates the direction of nucleophilic attack.
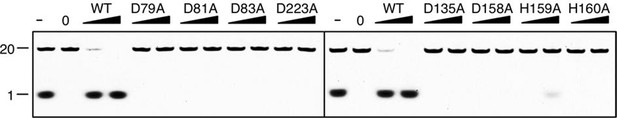
Denaturing PAGE gel showing the inactivation of mutant drRecJ proteins (alanine substitutions of residues involved in metal-ion-chelation).
KY08 was incubated with drRecJ proteins (0, 5 and 20 nM) in the presence of 100 nM Mn2+.
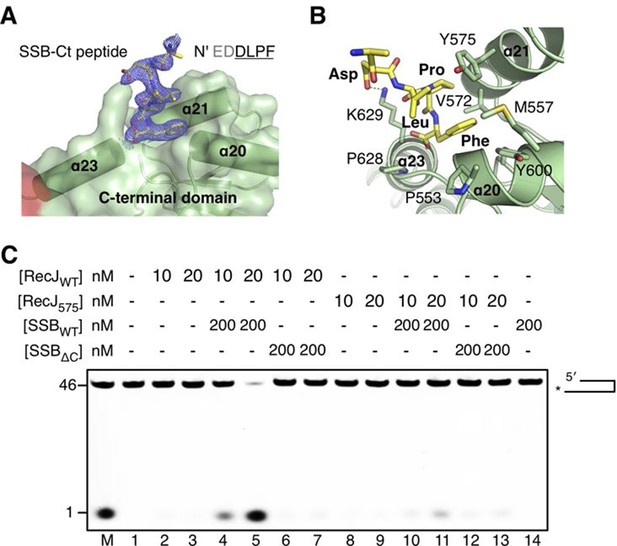
The C-terminal domain interacts with the SSB-Ct.
(A) SSB-Ct binding pocket. SSB-Ct is shown as stick. The electron density of SSB-Ct is shown in blue with the refined 2Fo-Fc map contoured at 1σ. (B) Interactions of the SSB-Ct and C-terminal domain. Both SSB-Ct (yellow) and residues involved in SSB-Ct interactions (green) are shown as sticks. The ionic bond between the Asp298 of SSB-Ct and Lys629 of drRecJ is indicated by a yellow dashed line. (C) SSB enhances wild-type drRecJ degradation. Y575A mutant drRecJ (RecJ575), wild-type drSSB (SSBWT) and drSSB lacking eight C-terminal residues (SSBΔC) were purified to perform the nuclease assays. For the reaction, DNA with a 3´-ssDNA overhang was pre-incubated with 200 nM SSBWT or SSBΔC and treated with drRecJ or RecJ575 (5 and 20 nM) (see Materials and methods).
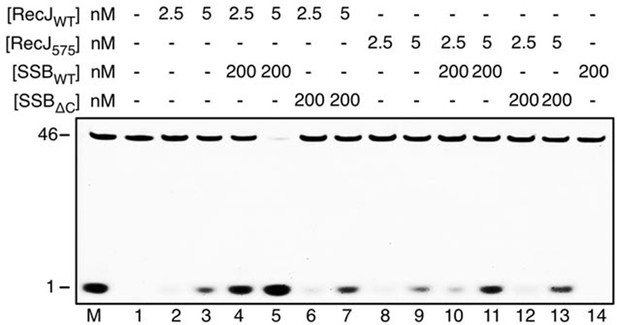
SSB enhances wild-type drRecJ degradation on ssDNA.
Y575A mutant drRecJ (RecJ575), wild-type drSSB (SSBWT) and drSSB lacking eight C-terminal residues (SSBΔC) were purified to perform the nuclease assays. For the reaction, 100 nM ssDNA was pre-incubated with 200 nM SSBWT or SSBΔC and treated with drRecJ or RecJ575 (2.5 and 5 nM, or 10 and 20 nM) (see Materials and methods).
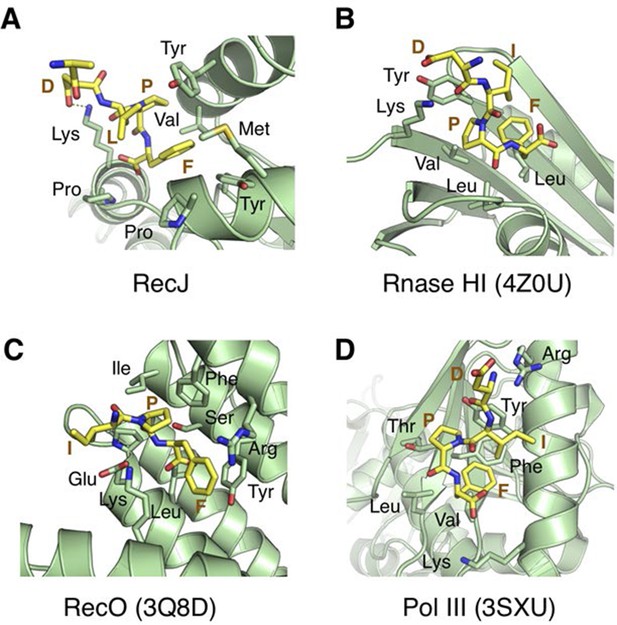
Structural comparison of the SSB-Ct binding pockets of RecJ, Rnase HI (4Z0U), RecO (3Q8D) and Pol III (3SXU).
The SSB-Ct are shown as sticks and colored in yellow. Residues interact with SSB-Ct are labeled, shown as sticks and colored in green.
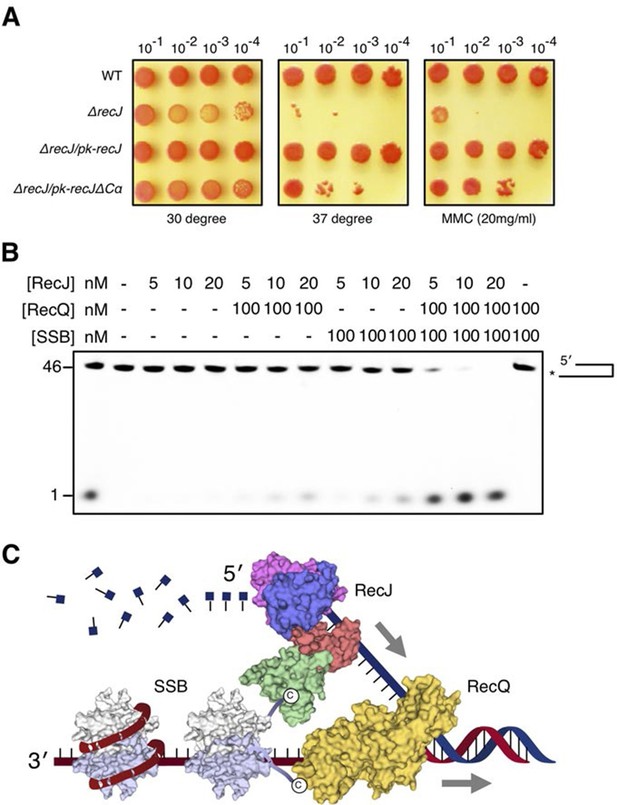
DSB end resection requires the coordinate activities of RecJ, RecQ and SSB proteins.
(A) Functional analysis of the drRecJ α-CT in vivo. Wild-type (WT), recJ mutant (ΔrecJ) and recJ complementary (ΔrecJ/pk-recJ for the entire RecJ and ΔrecJ/pk-recJΔCα for RecJ lacking α-CT) strains were spotted on TGY medium following high-temperature (37 degrees) and MMC treatments. (B) drRecJ processes DNA bearing 3´-ssDNA overhang together with drRecQ and drSSB. The reaction contained drRecJ (5, 10 and 20 nM), drRecQ (100 nM) and drSSB (100 nM). (C) A model for DSB end resection by RecJ, RecQ and SSB proteins in D. radiodurans. RecQ (yellow) is bound to the ss-dsDNA junction, which unwinds them to generate 5´-tailed ssDNA. Following RecJ digestion, the SSB (homodimer, white and grey) is recruited to the resultant 3´-ssDNA overhang, which facilitates further strand exchange reaction.
Tables
Statistics from crystallographic analysis.
| RecJ-dTMP | RecJd-DNA | RecJd-DNA-SSBct | |
|---|---|---|---|
| Complex I | Complex II | Complex III | |
| Data collection | |||
| Space group | P 3221 | P 3221 | P 3221 |
| Cell dimensions a, b, c (Å) | 106.53 | 105.83 | 102.22 |
| 106.53 | 105.82 | 102.22 | |
| 161.90 | 165.40 | 166.12 | |
| Wavelength (Å) | 0.9792 | 0.9792 | 0.9792 |
| Resolution (Å) | 30–2.7 (2.77–2.70) | 30–2.6 (2.66–2.60) | 30–2.3 (2.35–2.30) |
| R-meas | 5.5 (64.1) | 6.5 (79.1) | 7.4 (63.7) |
| I/σI | 27.0 (4.0) | 22.6 (3.0) | 17.0 (2.9) |
| Completeness (%) | 98.9 (99.5) | 99.6 (99.4) | 99.4 (94.1) |
| Redundancy | 8.9 | 8.3 | 7.2 |
| Refinement | |||
| Resolution (Å) | 30–2.7 | 30–2.6 | 30–2.3 |
| No. reflections | 29747 | 33594 | 45296 |
| Rwork/Rfree | 20.61/25.20 | 18.47/22.84 | 22.56/23.89 |
| No. atoms | |||
| Protein/DNA | 5262/- | 5373/286 | 5342/182 |
| Ligand/Ion | 21/2 | 35/2 | 55/2 |
| Waters | 12 | - | 142 |
| B factors | |||
| Protein/DNA | 79.8/- | 70.4/103.9 | 58.2/93.7 |
| Ligand/Ion | 66.7/76.6 | 99.8/68.2 | 74.5/61.9 |
| Water | 62.7 | - | 48.9 |
| Rmsd | |||
| Bond length (Å) | 0.012 | 0.005 | 0.012 |
| Bond Angle (°) | 1.085 | 0.849 | 1.464 |
| Ramachandran statitics | |||
| Favored (%) | 98.0 0 | 99.1 | 98.9 |
| Allowed (%) | 2.0 | 0.9 | 1.1 |
| Outliers (%) | 0 | 0 | 0 |
-
Values in parentheses refer to the highest resolution shell.
-
R factor = Σ||F(obs)- F(calc)||/Σ|F(obs)|.
-
Rfree = R factor calculated using 5.0% of the reflection data randomly chosen and omitted from the start of refinement.
-
RecJd denotes catalytic inactive drRecJ (H160A)
Kinetic parameters of wild-type and mutant drRecJ proteins.
| Protein-substrate | Km (nM) | kcat (min-1) | kcat/Km (µM-1 min-1) |
|---|---|---|---|
| WT-KY09 (poly(dT)) | 74.9 ± 7.6 | 1.61 ± 0.03 | 21.5 |
| WT-KY08 (poly(dA)) | 100.3 ± 5.5 | 1.15 ± 0.02 | 11.5 |
| Y496A -KY09 (poly(dT)) | 102.2 ± 10.9 | 0.32 ± 0.01 | 3.1 |
| Y496A -KY08 (poly(dA)) | 109.1 ± 8.4 | 0.27 ± 0.01 | 2.5 |
| WT-KY03 | 90.3 ± 4.5 | 1.55 ± 0.01 | 17.1 |
| Y80A-KY03 | 223.1 ± 12.2 | 0.25 ± 0.02 | 1.1 |
| Y114A-KY03 | 349.4 ± 23.8 | 0.77 ± 0.05 | 2.2 |
| R109A-KY03 | 158.3 ± 19.9 | 0.25 ± 0.02 | 1.6 |
| S371A-KY03 | 104.4 ± 8.8 | 0.78 ± 0.03 | 7.5 |
| R373A-KY03 | 349.6 ± 26.2 | 0.61 ± 0.03 | 1.7 |
| R393A-KY03 | 105.8 ± 6.9 | 1.51 ± 0.02 | 14.2 |
| H397A-KY03 | 321.3 ± 22.0 | 0.77 ± 0.03 | 2.4 |
Additional files
-
Supplementary file 1
Primers used for cloning and mutagenesis.
- https://doi.org/10.7554/eLife.14294.023
-
Supplementary file 2
Strains and plasmids used in this study.
- https://doi.org/10.7554/eLife.14294.024
-
Supplementary file 3
DNA substrates used in this study.
- https://doi.org/10.7554/eLife.14294.025





