Quorum sensing control of Type VI secretion factors restricts the proliferation of quorum-sensing mutants
Figures
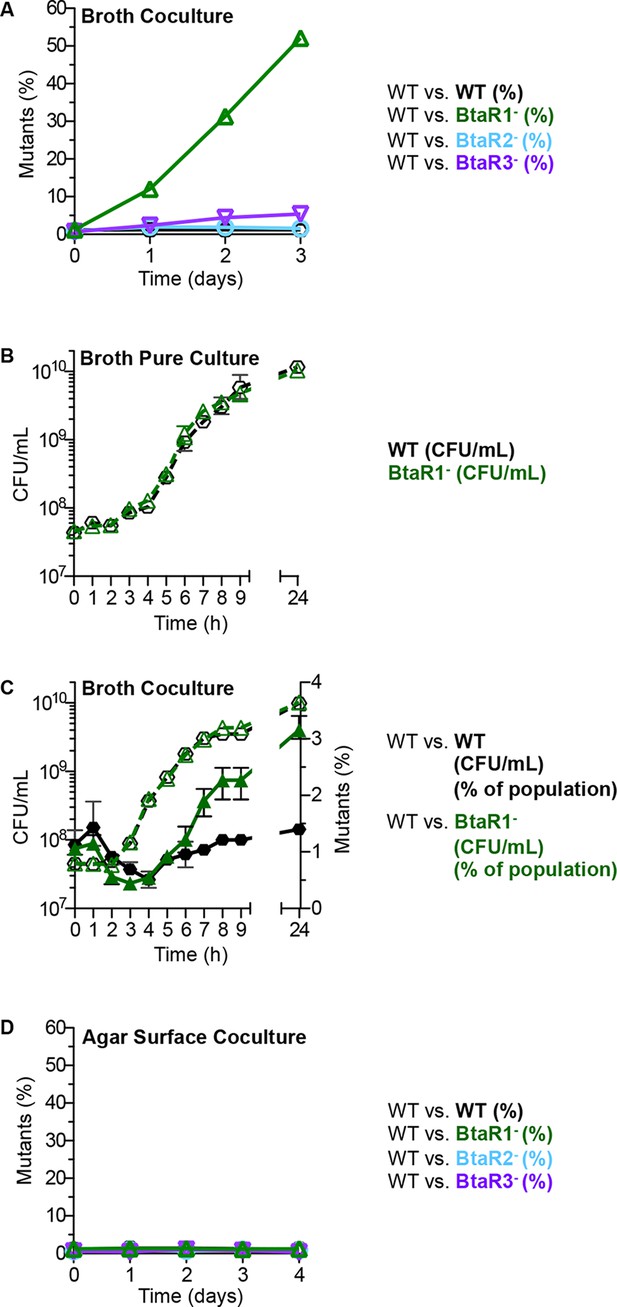
Growth of the wild type and QS mutants in LB broth and on an agar surface.
(A) In LB broth, the wild type (strain E264) was mixed with the BtaR1- mutant CM157, the BtaR2- mutant CM159 or the BtaR3- mutant CM161 at a starting mutant relative abundance of 1%. All mutants were KmR. As a control we mixed the wild type with CM218, a KmR strain derived from the wild type. Results are percent of total colonies that are KmR (% mutants). The control shows there is little cost associated with the KmR marker. (B) LB broth pure culture growth curves of CM224, a TpR wild-type strain and the KmR BtaR1- mutant. Data for Figure 1B for the statistical analysis described in the text are in the Figure 1—source data 1). (C) 24-hour LB broth coculture. The TpR marked wild type) mixed with the KmR marked wild type), or the BtaR1 mutant. KmR bacteria were started at 1% of the population and each coculture was grown in a flask for 24 hr. Bacteria were enumerated by colony counts for total coculture yield (CFU/mL) and percent KmR mutants in the population (open symbols are CFU/mL, closed symbols are percent of mutants). Bars show the mean and range of at least three biological replicates. (D) Agar surface competitions. Wild type with the BtaR1- mutant, the BtaR2- mutant or the BtaR3- mutant at a starting mutant abundance of 1%. As a control we mixed the wild type with the KmR strain derived from the wild type. Data are means of at least three biological replicates. Some ranges are not visible, as they were small and within the size of the symbols.
-
Figure 1—source data 1
Experimental data for the statistical analysis for Figure 1B.
- https://doi.org/10.7554/eLife.14712.004
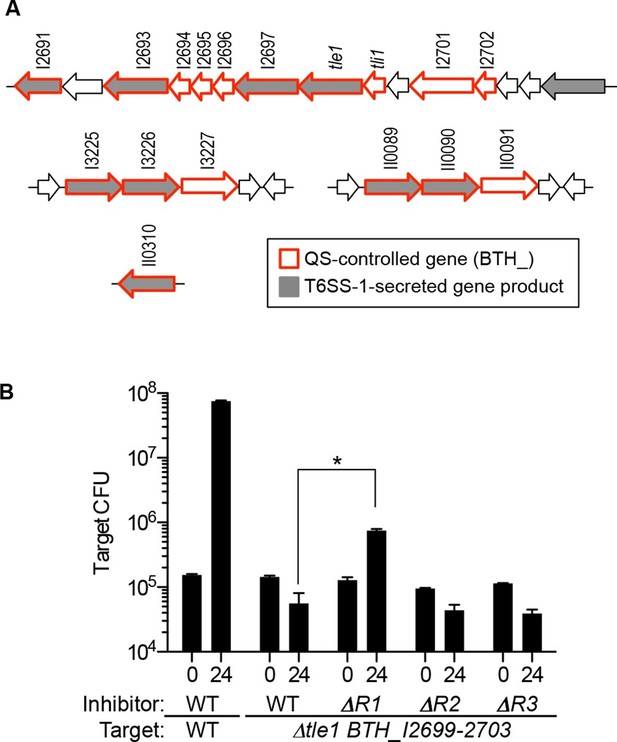
Map of QS-controlled genes for T6SS-1 substrates and solid-surface competition experiments with T6SS-1 mutants.
(A) Genes coding for known or putative T6SS-1 substrates (Russell et al., 2012) are shaded grey and genes controlled by QS (Majerczyk et al., 2014) are outlined in red. BTH_I2698 is the tle1 toxin gene and BTH_I2699 is the tli1 immunity gene (Russell et al., 2013). The neighboring genes, BTH_I2701 and BTH_I2702 are paralogs of tle1 and tli1. (B). Agar surface competition experiments. These experiments were started with a relative abundance of 10% target, CM316 TpR T6SS-1 immunity mutant (△tle1△BTH_I2699-703) or CM218 TpR wild type (WT). The inhibitor strains all carried a KmR marker; The wild type inhibitor was CM224, The BtaR1- mutant (△R1) was CM157, the BtaR2- mutant (△R2) was CM159, and the BtaR3- mutant (△R3) was CM161. Target cell abundance after one 24 hr round of colony growth was determined by plate counting. Data are from three biological replicates. The asterisk (*) indicates statistical significance between the CFU/mL measured in the WT or BtaR1 culture, as determined by an unpaired t test. Data can be found in the Figure 2—source data 1.
-
Figure 2—source data 1
Experimental data for statistical analysis of the results in Figure 2.
- https://doi.org/10.7554/eLife.14712.006
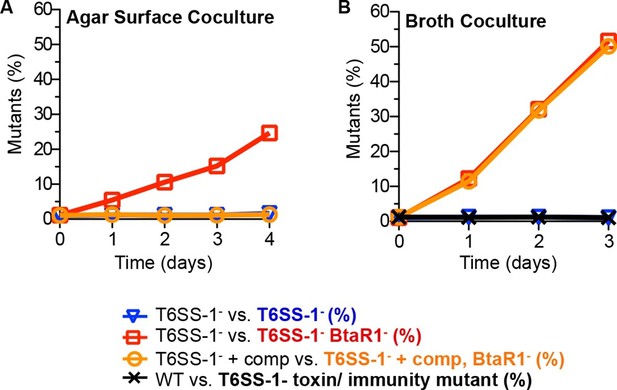
The contributions of T6SS-1 in policing BtaR1- mutants.
Competitions were on (A) LB agar plates for solid surface cocultures or in (B) LB broth for liquid cocultures. The T6SS-1-mutant (△clpV) was mixed with the TpR BtaR1- T6SS-1- mutant ES9 (open red squares) or the KmR T6SS-1- mutant ES18 (open blue downward triangle). The clpV complemented T6SS-1- mutant △clpV-comp was mixed with the clpV complemented BtaR1- T6SS-1- mutant ES16 (open orange circles). For liquid cultures, the wild type (strain E264) was mixed with CM316 Tpr T6SS-1 toxin/ immunity mutant (△tle1△BTH_I2699-703) (black crosshatch). This served as a control to demonstrate T6SS-1-mediated killing does not occur in liquid broth cocultures. Results are the mean of at least three biological replicates. Ranges were within the size of the symbols.
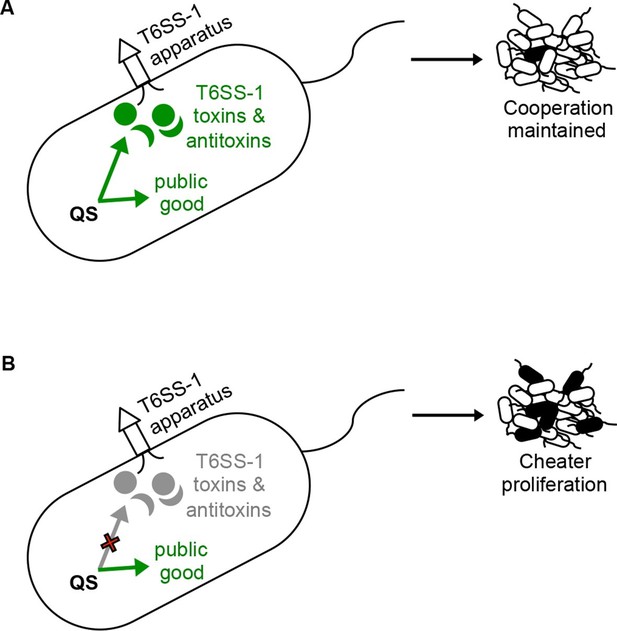
A policing model where QS control of T6S contact-dependent killing (effector and immunity proteins) enforces cooperation.
(A) The bacterium on the left depicts a situation where QS co-regulates a public good with the T6SS-1 effector and immunity factors. Under these conditions, QS-proficient bacteria (cooperators, shown as white cells) limit the proliferation of a QS mutant (potential cheater, black cell) in a population. (B) In this situation, QS no longer co-regulates public good production with the T6SS-1 killing mechanism. Without co-regulation of the killing mechanism, QS mutants (potential cheaters, black cells) outcompete the wild type cooperators (white cells).
Tables
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Genotype or descriptiona | Source | |
|---|---|---|---|
| Bacterial strains | |||
| DH10B | E. coli cloning vehicle | Invitrogen | |
| E264 | Wild-type B. thailandensis | (Brett et al., 1998) | |
| JBT107 | E264 △btaR1 | (Chandler et al., 2009) | |
| JBT108 | E264 △btaR2 | (Chandler et al., 2009) | |
| JBT109 | E264 △btaR3 | (Chandler et al., 2009) | |
| CM157 | JBT107 glmS1 attn7::Km; KmR | This study | |
| CM159 | JBT108 glmS1 attn7::Km; KmR | This study | |
| CM161 | JBT109 glmS1 attn7::Km; KmR | This study | |
| CM218 | E264 glmS1 attn7::Km; KmR | This study | |
| CM224 | E264 glmS1 attn7::Tp; TpR | This study | |
| BT03399 | E264 btaR1139::ISlacZ/PrhaBo-Tp/FRT; TpR | (Gallagher et al., 2013) | |
| △clpV | E264 △BTH_I2958 | (Schwarz et al., 2010) | |
| △clpV-comp | △clpV att Tn7-miniTn7T-Tp-S12-BTH_I2958 | (Schwarz et al., 2010) | |
| ES9 | △clpV btaR1139::ISlacZ/PrhaBo-Tp/FRT; TpR | This study | |
| ES18 | △clpV glmS1 attn7::Tp; TpR | This study | |
| ES14 | △clpV btaR1139::ISlacZ/PrhaBo scar; TpS | This study | |
| ES15 | ES14 glmS1 attn7::PS12-clpV, Tp; TpR | This study | |
| ES16 | ES15 glmS2 attn7::Km; TpR, KmR | This Study | |
| △tle1△BTH_I2699-703 | E264 △BTH_I2698-BTH_I2703 | (Russell et al., 2013) | |
| CM316 | △tle1 △12699-2700 glmS1 attn7::Tp; TpR | This study | |
| Plasmids | |||
| pTNS2 | R6K replicon TnsABC+D vector | (Choi et al., 2005) | |
| pUC18T-mini-Tn7T-Tp | Cloning vector | (Choi and Schweizer, 2006) | |
| pUC18T-mini-Tn7T-Km-FRT | Cloning vector | (Choi et al., 2008) | |
| pFLPe2 | Flp recombinase-expressing vector; ZeoR | (Choi et al., 2008) | |
| pUC18T-miniTn7T-Tp-S12-BTH_I2958 | clpV complementation contruct (PS12-clpV) | (Schwarz et al., 2010) | |





