QIL1 mutation causes MICOS disassembly and early onset fatal mitochondrial encephalopathy with liver disease
Figures
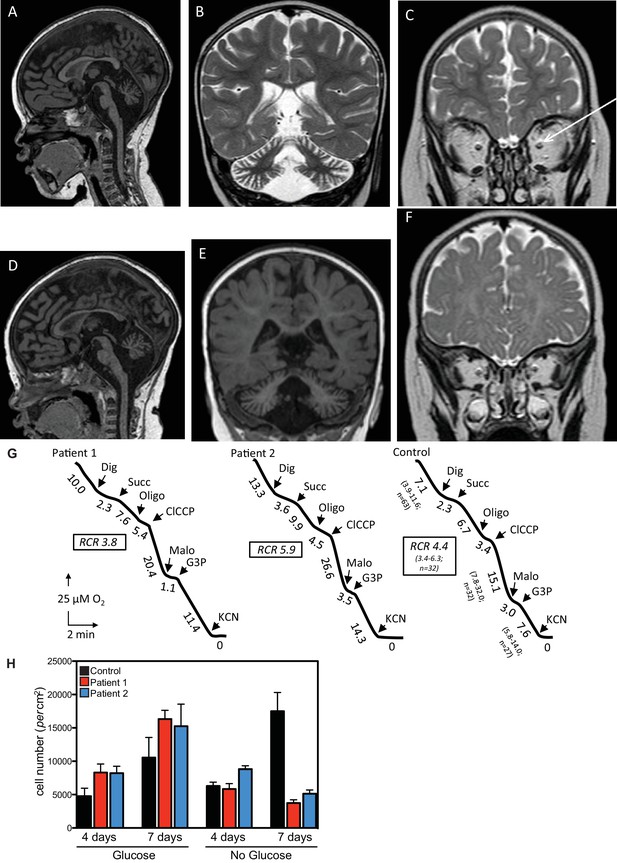
Clinical presentation of patients deficient in QIL1.
(A–C) Brain MRI of patients 1 (A–C, 2 years of age) and 2 (D–F, 1 year of age). T1-weighted sagittal views show cerebellar atrophy of the vermis and the brainstem (A,D). T2-weighted (B, patient 1) and FLAIR (E, patient 2) coronal views show cerebellar hemisphere atrophy. T2-weighted coronal views (C,F) show optic atrophy (arrow, patient 1). (G) Oxygen consumption by patients’ and control skin fibroblasts was measured as reported elsewhere (El-Khoury et al., 2013; Rustin et al., 1994). Cellular respiration was first measured using intact fibroblasts. It was essentially abolished upon addition of a limited amount of digitonin (Dig) which caused the leakage of endogenous respiratory substrates in the assay medium. Oxygen consumption resumed upon subsequent addition of 10 mM succinate (Succ) to the digitonin-permeabilized fibroblast. This oxidation decreased in the presence of oligomycin (oligo) (a mitochondrial ATPase inhibitor) while adding an uncoupling agent (m-ClCCP) allowed a maximal rate of oxidation and to calculate a respiratory chain control (RCR) value (rate in the presence of uncoupler versus rate in the presence of the ATPase inhibitor). Malonate (Malo) (a specific succinate dehydrogenase inhibitor) addition essentially abolished oxygen uptake linked to succinate oxidation. Adding glycerol-3 phosphate (G3P) allowed the oxygen uptake to resume, this latter being fully inhibited by the addition of cyanide (KCN). The values along the traces are nmol/min/mg protein. (H) Control (black bars) or patient (blue or red bars) fibroblasts were cultured in the presence and absence of glucose for 7 days and cell number determined after 4 and 7 days (n = 3).
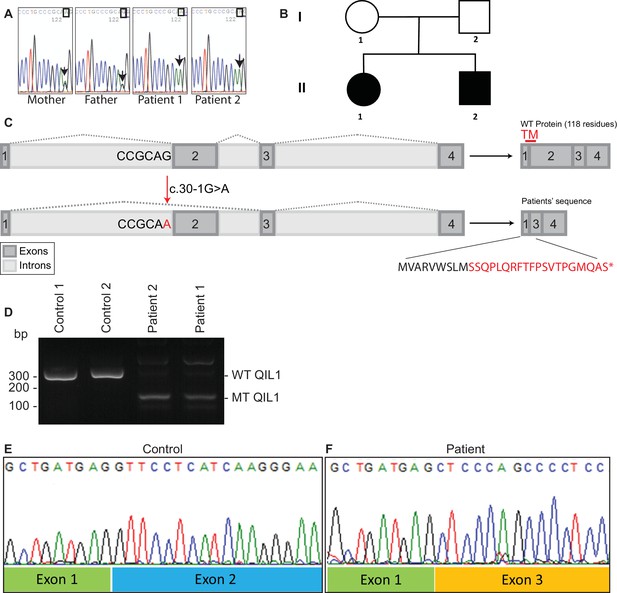
Identification of mutations in QIL1.
(A) Chromatogram depicting a homozygous mutation (c.30-1G>A) in C19orf70 (QIL1). (B) Pedigree showing autosomal recessive transmission. (C) Schematic illustration of the location of the mutation within the QIL1 transcript and the predicted consequences on splicing and the respective coding sequences. TM (red), indicates the position of a transmembrane domain in QIL1. (D) Ethidium bromide stained agarose gel of RT–PCR QIL1 products from controls and patients 1 and 2. (E–F) Sequence analysis of control (E) and patient 1 (F) QIL1 cDNAs.
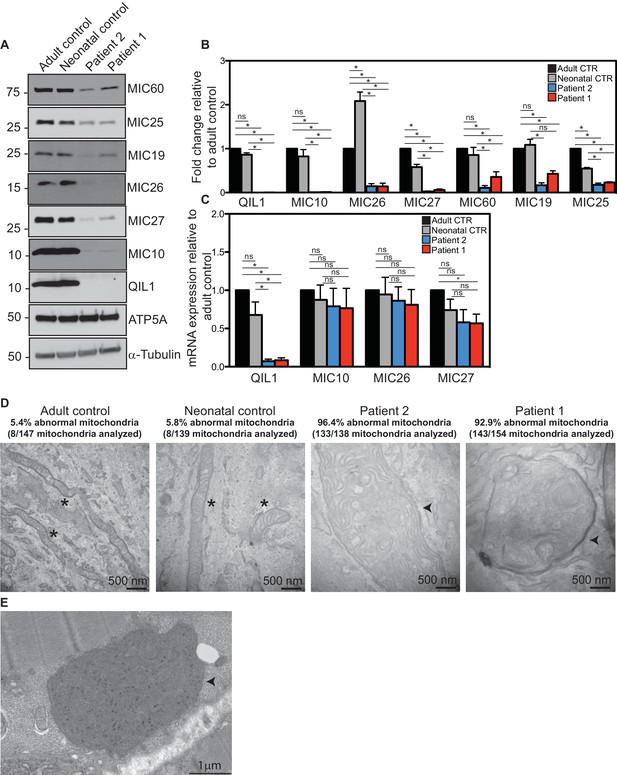
Reduced MICOS subunit abundance and cristae morphology defects in QIL1-deficient patients’ fibroblasts.
(A) Immunoblot analysis of QIL1 and various MICOS subunits in control adult skin fibroblasts, control neonatal skin fibroblasts and skin fibroblasts obtained from patients 1 and 2. Anti-ATP5A and α-Tubulin were used as loading controls. (B) Densitometry analysis was performed using ImageJ. Values were normalized to ATP5A. (C) qPCR analysis. Expression levels were normalized to Tubulin. (D) Electron microscopy analysis of control adult skin fibroblasts, control neonatal skin fibroblasts and skin fibroblasts from patients 1 and 2 showing enlarged mitochondria with cristae membrane swirls and proliferation of inner membranes in cells from both patients, as compared to normal mitochondria in control cells; some patients’ mitochondria contain electron dense inclusions. Morphologically abnormal mitochondria are indicated by the arrowhead. Mitochondria with cristae junctions of normal morphology are indicated with an asterisk. Quantification of abnormal mitochondria based on analysis of the indicated number of mitochondria by electron microscopy is shown. (E) Electron microscopy analysis of skeletal muscle biopsy from patient 1 showing large round mitochondria (arrowhead), which can sometimes reach the size of two sarcomeres. These large mitochondria show an important proliferation of membranes; some mitochondria contain inclusions in the form of electron dense dots. For panels B and C, asterisks represent p values<0.05. Error bars (± SEM) show the mean of 3 or 4 biological replicates.
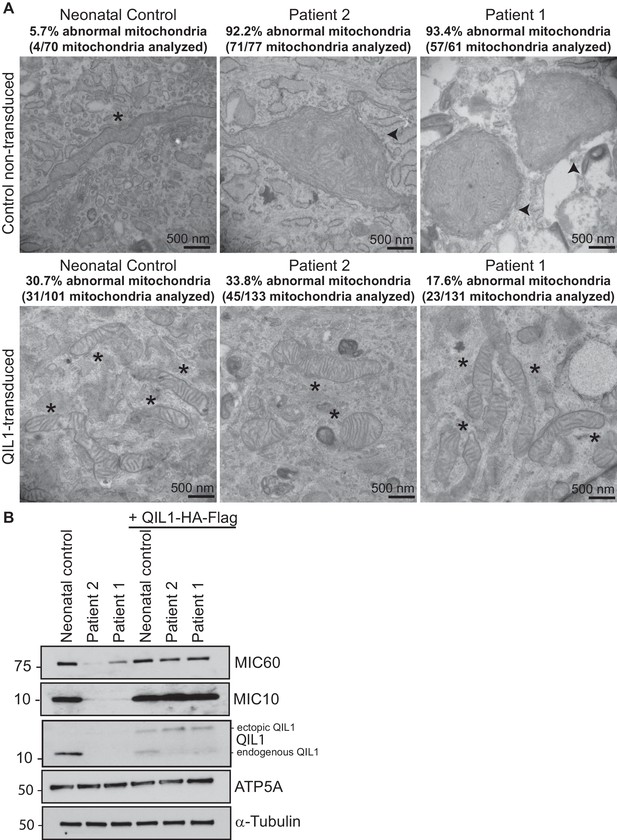
Ectopic QIL1 expression rescues mitochondrial morphological defects in patients’ fibroblasts.
(A) Electron micrographs of control neonatal skin fibroblasts and skin fibroblasts from patients 1 and 2 showing the rescue of mitochondria cristae morphology and shape upon ectopic QIL1-HA-Flag expression. Morphologically abnormal mitochondria are indicated by the arrowhead. Mitochondria with cristae junctions of normal morphology are indicated with an asterisk. Quantification of abnormal mitochondria based on analysis of the indicated number of mitochondria by electron microscopy is shown. (B) Immunoblot analysis of MICOS subunits MIC10 and MIC60 in control versus patients’ skin fibroblasts with or without overexpression of C-terminally HA-Flag tagged QIL1 demonstrating the rescue of the abundance of MICOS subunits upon ectopic QIL1 expression.
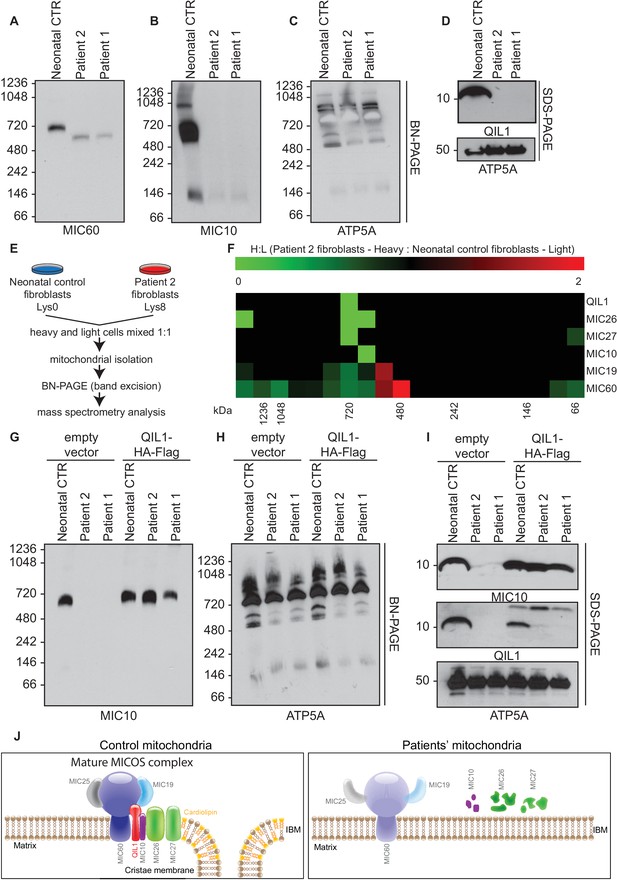
Patients’ mitochondria display MICOS assembly defects.
(A–C) Blue native electrophoresis followed by immunoblot analysis of MIC60, MIC10 and ATP5A in control neonatal and patients’ mitochondria from skin fibroblasts. In Panel D, mitochondrial extracts were subjected to SDS-PAGE and immunoblotting with the indicated antibodies. (E,F) Cells from control neonatal fibroblasts were labeled with light lysine (K0) while fibroblasts obtained from patient 2 were labelled with heavy lysine (K8). Cells were mixed at a 1:1 ratio and mitochondria were subsequently isolated, lysed in 1% digitonin and native protein complexes were separated in a blue native gel. Gel bands from >1 MDa to ~66 kDa were excised and subjected to mass spectrometry analysis. Heavy: Light ratios were calculated for the sum of all peptides quantified for each MICOS subunit and represented in a heatmap where ratios below 1 are represented in green and ratios above 1 are represented in red. (G,H) Blue-native gel analysis of MIC10 and ATP5A containing complexes from neonatal control fibroblasts or fibroblasts from patients 1 and 2 with or without rescue by stable expression of QIL1-HA-FLAG. Panel G, anti-MIC10. Panel H, anti-ATP5A. (I) Mitochondrial lysates from panels G–H were examined by SDS-PAGE and immunoblotting using the indicated antibodies. (J) Schematic representation of the effect of QIL1 loss on MICOS assembly in patients’ fibroblasts. In the mature MICOS complex in control cells, multiple transmembrane components of MICOS (MIC60, MIC10, MIC26, MIC27 and QIL1) associate with MIC25 and MIC19 to promote the formation of cristae membrane structures. In the mitochondria of patient cells lacking QIL1, the abundance of MIC10, MIC26, and MIC27 is greatly reduced, leading to loss of the MICOS complex and the absence of normal cristae structures within mitochondria. Blue-native gel analysis of MICOS from QIL1-deficient cells revealed a ~500 kDa complex containing reduced levels of MIC60 and MIC19, which appears to be unable to maintain cristae structure morphology within mitochondria.
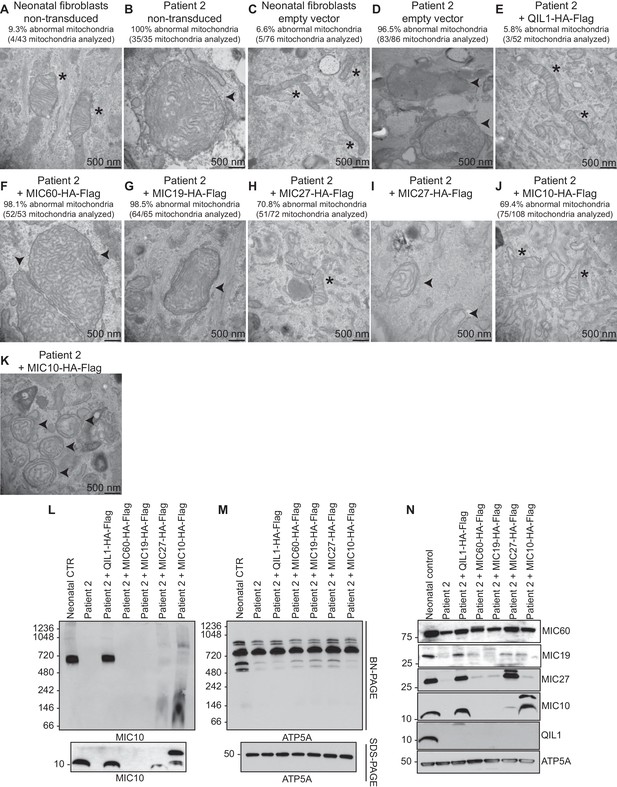
Analysis of QIL1-mutant fibroblasts ectopically expressing MICOS complex subunits.
(A–K) The indicated parental cell lines were transduced with the indicated lentivirus expressing a C-terminally HA-FLAG tagged MICOS subunit and cells subjected to electron microscopy. Morphologically abnormal mitochondria are indicated by the arrowhead. Mitochondria with cristae of normal morphology are indicated with an asterisk. Quantification of mitochondrial morphology in patient fibroblasts in panels A–K is shown. (L,M) Blue-native gel analysis of mitochondrial lysates from the indicated patient fibroblasts expressing the indicated MICOS subunits and corresponding SDS-PAGE analysis. Blots were probed with anti-MIC10 to examine the MICOS complex and with anti-ATP5A as a control. (N) Whole cell extracts from the cells employed in Panels L–M were subjected to SDS-PAGE and probed with the indicated antibodies.
Additional files
-
Supplementary file 1
Clinical and laboratory findings and respiratory chain activities in control and QIL1-defective patients.
(A) Clinical and laboratory findings from patients 1 and 2. Nd: non determined; H: hours of life; mo: months of life. 3MGCA: 3-methylglutaconic aciduria. (B) Respiratory chain activities in control and QIL1-defective patients’ fibroblasts. In fibroblasts, patients’ respiratory chain complexes activities were in control ranges. Complex IV activity normalized to citrate synthase was however more than 3 standard deviations lower than control, indicative a partial defect in complex IV. Activities are expressed in nmol/min/mg protein. (C) Respiratory chain activities in control and QIL1-defective patients’ skeletal muscle for patient 1. In muscle, all respiratory chain complexes (I-IV) activities normalized to citrate synthase were defective. Activities are expressed in nmol/min/mg protein.
- https://doi.org/10.7554/eLife.17163.008
-
Supplementary file 2
Primer sequences for QIL1 genomic DNA (gDNA) and complementary DNA (cDNA).
Tables show QIL1 primers used for PCR amplification.
- https://doi.org/10.7554/eLife.17163.009





