Nonenzymatic copying of RNA templates containing all four letters is catalyzed by activated oligonucleotides
Figures
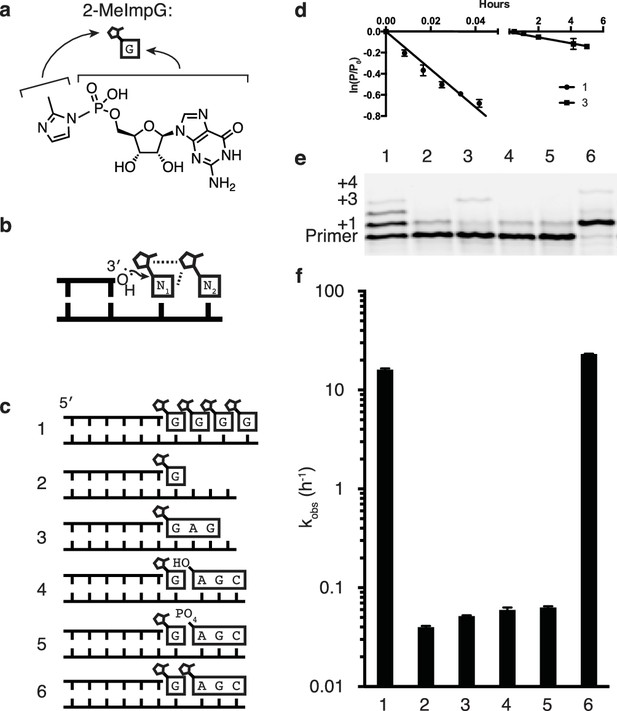
Catalysis of nonenzymatic primer extension by activated downstream nucleotides.
(a) Structure of 2-methylimidazole-activated guanosine-5′-monophosphate, and its schematic representation. (b) Schematic of the RNA primer extension reaction. N1 represents an individual ribonucleotide monomer in position to react with the primer, N2 represents either a monomer or an oligomer downstream, with a leaving group capable of interacting with N1. Dashed lines: Potential interactions between leaving groups or between the downstream leaving group and the upstream nucleotide N1. (c) Schematics of the RNA primers, templates, monomers and oligomers used in d–f. Templates are complementary to the displayed monomers and oligomers. Template 2 has a U following the C to which the G monomer is bound to prevent downstream binding of G. (d) Primer extension by polymerization or ligation on templates 1 or 3, respectively. Fits describe ln(fraction primer remaining) vs time, giving an apparent first-order rate constant (Figure 1—figure supplement 1). (e) Primer extension assay for the experiments described in c, showing reaction progress after 10 min. In lane 6, a primer + 4 band can be observed, representing the slow addition of the activated trimer after the monomer. (f) Pseudo-first order rates of the reactions described in c. Error bars indicate S.E.M; all experiments were performed in triplicate or greater. Reaction conditions: 10 μM primer, 11 μM template, 200 mM CHES pH 9.0, 200 mM MgCl2, 50 mM monomer, 1 mM trimer.
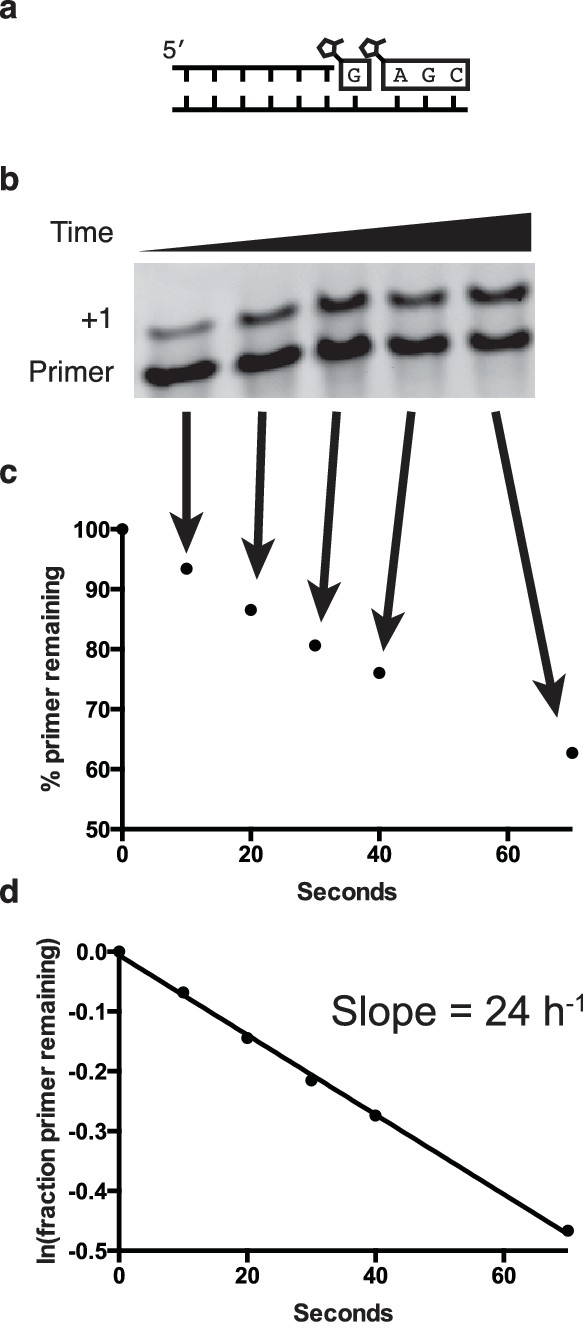
Determination of the rates of primer extension reactions.
(a) Schematic of a primer extension reaction. (b) Gel showing primer (starting material) and extended primer. (c) Plot showing the decay of primer as a percentage of total fluorescence intensity on the gel. (d) Following the assumption of a pseudo-first order rate curve, the ln(fraction of primer remaining) plotted over time gives a plot where the slope is the rate for the reaction.
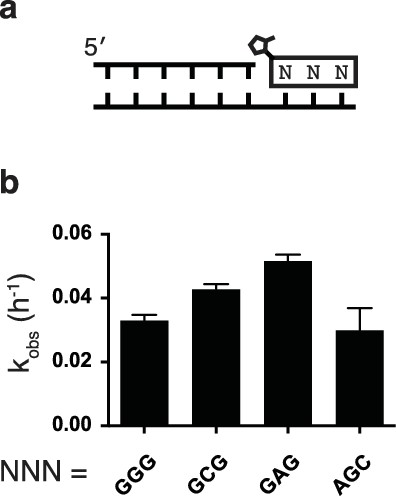
Rates of additional trimer ligations.
(a) Schematic of trimer ligation reaction. (b) Calculated rates of ligation of various trimer sequences.
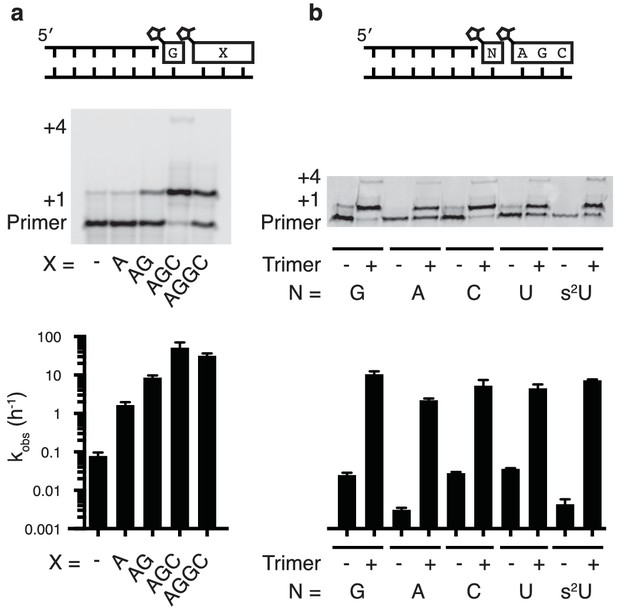
Nonenzymatic primer extension using all four monomers.
(a) RNA primer extension assay using alternative 'helper' oligomers and corresponding, complementary templates. The respective rates are shown at bottom. Concentration curves are provided for AG and AGC in Figure 2—figure supplement 1. (b) Primer extension assay for each of the four monomers, as well as 2-thiouridine, in the absence and presence of activated trimer. Shown at bottom of both a and b is a bar graph with the respective rates. Reaction conditions: 10 μM primer, 11 μM template, 200 mM Tris pH 8.0, 100 mM MgCl2, 50 mM monomer, 10 mM 2-MeImpAG, 1 mM 2-MeImpAGC, 1 mM 2-MeImpAGGC. The gels in a and b show reaction progress after 10 min and 1 hr respectively. Error bars indicate S.E.M; all experiments were performed in triplicate or greater.
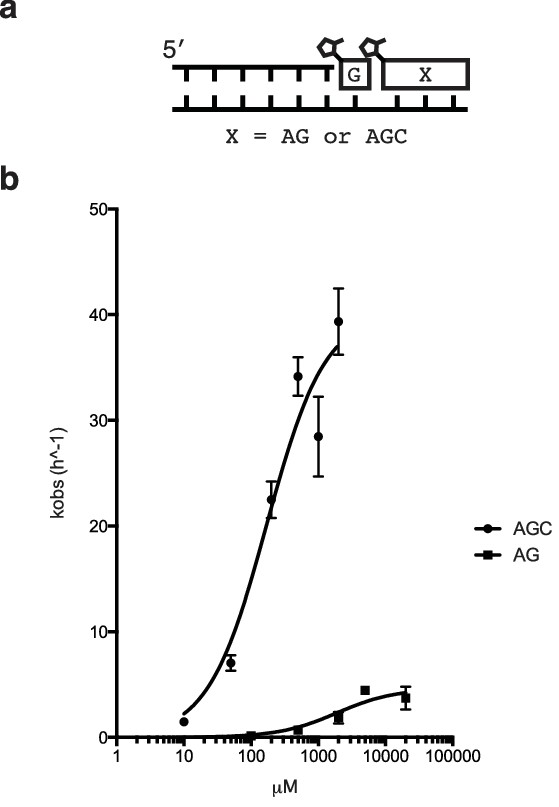
Rate of primer extension vs. concentration of downstream activated di- and tri-nucleotides.
(a) Schematic of the primer extension reaction. (b) Saturation curves of primer extension reactions with either AG or AGC as the helper oligomers.
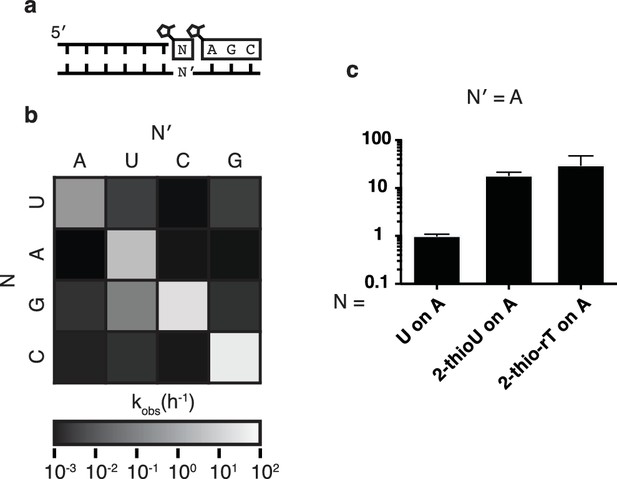
Fidelity of trimer-assisted primer extension.
(a) Schematic of the fidelity assay. Using the same RNA primer and trimer, four different templates, one with each base, were paired with each of the four 2-methylimidazole activated monomers to test the relative rates of each matched and mismatched pair. (b) Heat map showing the relative rates of primer extension for each template:monomer pairing. Black indicates a rate of 0.001 h−1, white indicates a rate of 100 h−1. A bar graph and a table of the same data can be found in Figure 3—figure supplement 1. (c) Relative rates of primer extension with the modified U monomers, 2-thiouridine and 2-thioribothymidine compared to that of a canonical, activated U monomer on a template containing an A. Reaction conditions as in Figure 2. Error bars indicate S.E.M. All experiments were performed in triplicate or greater.
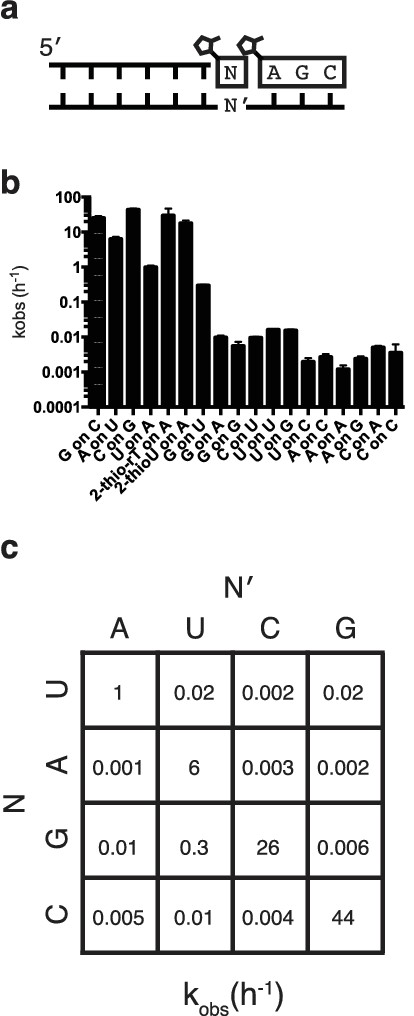
Rates of trimer-assisted primer extension for all monomer/template combinations.
(a) Schematic of the primer extension reaction. (b) Rates of monomer additions on different templates for all monomer-template pairs. (c) The rates from part b on the grid from Figure 3b.
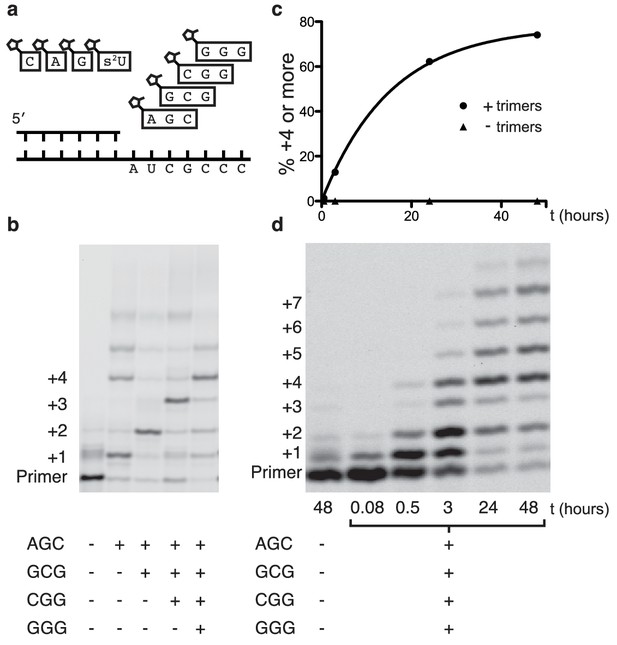
Primer extension with all four monomers in a one-pot reaction.
(a) Schematic of a primer extension reaction incorporating all four RNA monomers, with four respective downstream helper trimers. (b) All four monomers and the activated trimers indicated below the gel were added to each reaction. For example, in lane 3 all four monomers and the activated AGC and GCG trimers were mixed with primer, template, buffer and Mg2+. Higher bands represent either the ligation of activated trimers or the polymerization of G monomers without trimer assistance. The gel shows reaction progress after 16 hr. (c) Timecourse of the reaction with all four trimers. A 48 hr timepoint of the reaction without any trimers (left lane of b) is contrasted with a timecourse (timepoints at 5 and 30 min, 2, 24 and 48 hr). The percentage of polymerization products that have been extended by at least four nucleotides is plotted over time. (d) A gel displaying the timepoints plotted in part c. A timecourse showing the effect of overlapping trimers on reaction rate can be found in Figure 4—figure supplement 1. Reaction conditions: 2.5 μM primer, 2.5 μM template, 100 mM HEPES pH 8.0, 100 mM MgCl2, 20 mM monomer, 100 μM trimer.
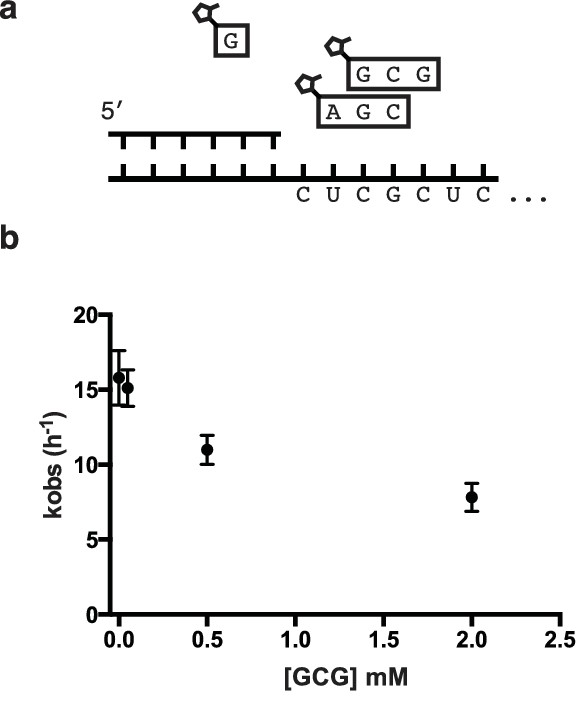
Inhibition of primer extension by overlapping helper-trimers.
(a) Schematic of the primer extension reaction. (b) Rate of primer extension as a function of increasing GCG trimer concentration.
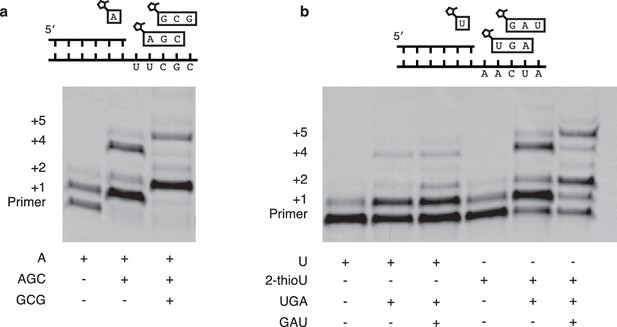
Extension of primers by multiple consecutive A or U nucleotides.
(a) Schematic of a primer extension reaction wherein two consecutive A nucleotides can polymerize. The gel shows extension without trimers and with one or two trimers after 16 hr. (b) Same as in (a) but with two U additions. In the right half 2-MeImpU is replaced with 2-MeImp-2-thioU.
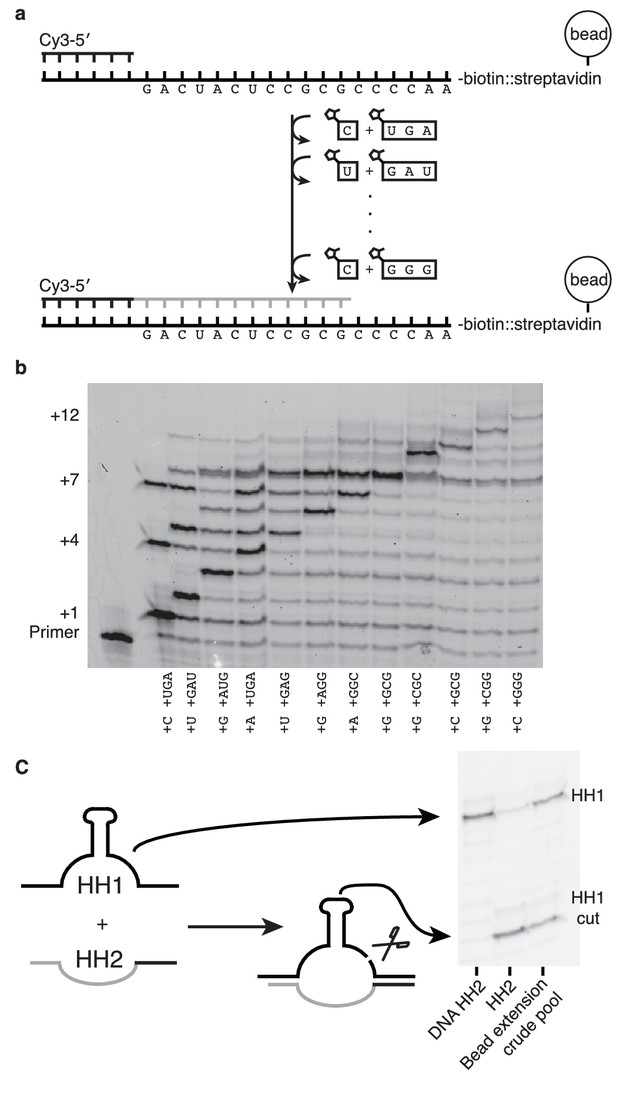
Nonenzymatic RNA synthesis of an active hammerhead ribozyme by template dependent primer extension.
(a) Schematic of the bead-assisted primer extension reaction. Synthesis of the hammerhead ribozyme strand HH2 by the sequential addition of matched pairs of activated monomers and trimers. (b) Each lane shows primer extension by one nucleotide, catalyzed by the corresponding downstream activated trimer. Final yield of the +12 product was 6%. Reaction conditions: 100 mM MgCl2, 200 mM Tris-HCl pH 8, 25 mM monomer and 2 mM trimer. The beads were washed between steps in 2 M NaCl, 1 mM EDTA and 10 mM Tris-HCl pH 7. For steps 2, 5 and 7, 50 mM monomer and 4 mM trimer were added. Each step was monitored by gel electrophoresis until it reached >75% yield. Steps varied in length from 30 min to 22 hr; the total time required was 150 hr. (c) The product of the last bead-assisted reaction was removed from the bead by the addition of the an excess of the DNA sequence of the extended primer at 95°C over 2 min. After DNase treatment, the target hammerhead RNA, HH1, was added. The third lane shows the extent of the nuclease reaction, 50%, after 16 hr at room temperature. For comparison, pure HH2 made from DNA and RNA served as negative and positive controls respectively. Reaction conditions: Extended primer from the beads was added to 200 mM MgCl2, 200 mM Tris-HCl pH 8 and 250 nM Cy5 labeled HH1 RNA.
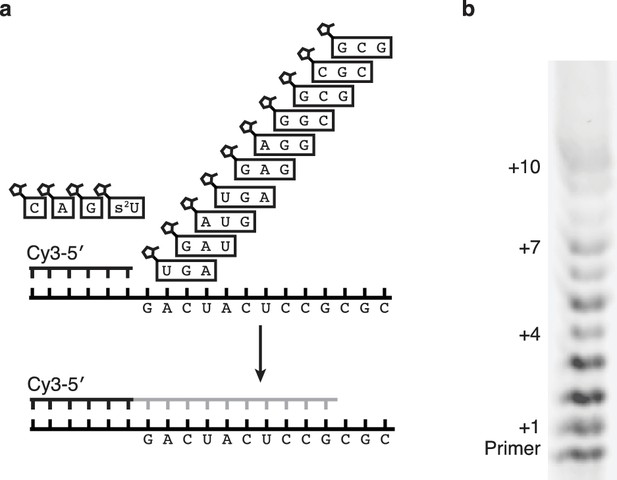
One pot synthesis of hammerhead ribozyme.
(a) Schematic of the primer extension reaction. (b) Gel showing primer extension after 18 hr. Reaction conditions: 5 μM primer, 5.5 μM template, 100 mM Tris pH 8, 100 mM MgCl2, 12.5 mM monomers, 500 μM trimers.





