RETRACTED: The structure of SV40 large T hexameric helicase in complex with AT-rich origin DNA
Figures
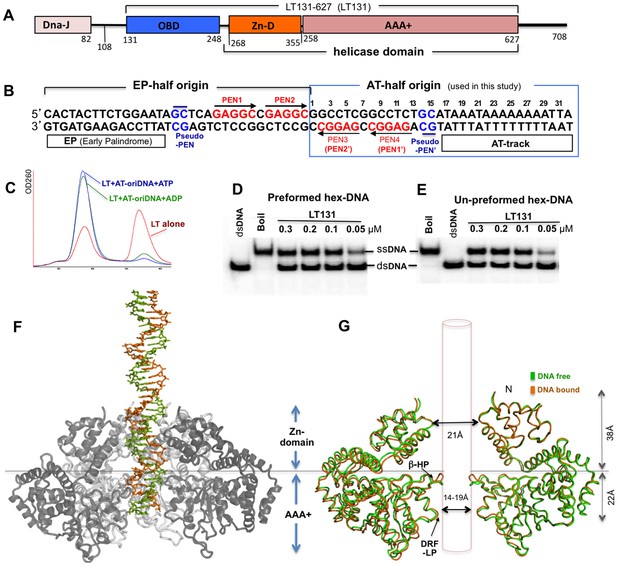
SV40 LT domain structure, core origin DNA (oriDNA), and the co-crystal structure of the LT helicase domain bound to the AT-half oriDNA.
(A) LT domain organization (drawn to scale), with the LT131 used for the co-crystallization study indicated at the top. (B) The full 64-bp core replication oriDNA of SV40. The four penta-nucleotides (PEN) (GAGGC (in red)) are flanked in the AT-half and in the EP-half origin. The AT-half origin DNA (boxed in blue) is co-crystallized with the LT hexameric helicase in this study. (C) The gel filtration profile (Superose 6) of LT131 alone or in complex with the AT-half oriDNA in the presence of ADP or ATP without Mg2+. LT131 alone equilibrates between hexamer and monomer peaks (red), but will form a stable hexamer when ATP is present (Gai et al., 2004a; Li et al., 2003). In the presence of ATP (blue) or ADP (green), LT131 forms stable hexamer-DNA complexes with the 32-bp AT-half oriDNA, which migrate slightly larger than the LT131 hexamer alone. The LT-oriDNA-ATP complex isolated from this peak showed no obvious dissociation after up to six hours, suggesting a very stable complex without ATP hydrolysis. (D, E) The preformed LT hexamer-oriDNA complexes (made by first incubating LT131 with oriDNA and ATP, then adding Mg2+ last to initiate unwinding, see Materials and methods) is active in unwinding the oriDNA (D). For comparison, (E) shows the unwinding results obtained using LT131 with no preformed (un-preformed) hexamer-oriDNA complexes (made by adding LT131 and Mg2+ last to initiate unwinding, see Materials and methods);this shows unwinding efficiency similar to that obtained using the preformed LT-oriDNA complex (D). This result suggests that the preformed stable LT-oriDNA complex is active in unwinding the oriDNA. Note that the unwound ssDNA product migrates slower than the dsDNA substrate because of hairpin formation at the palindrome origin sequence. (F) Overall view of the LT hexameric helicase bound to the AT-half origin DNA (32 bp) in the central channel. (G) Superimposition of the DNA-free (green) and DNA-bound (orange) hexamer structures, both of which are in an ADP-bound state. For clarity, only two subunits of hexamer are shown so that it is possible to visualize the β-hairpins and DRF loops. The Zn-domain and the AAA+ domain of LT are indicated.
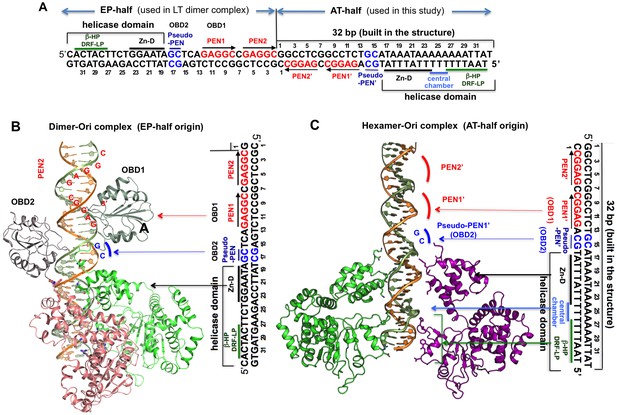
Comparing the DNA register in the dimeric LT structure bound to the EP-half origin with that in the hexameric LT structure bound to the AT-half origin DNA.
(A) The 64-bp full core origin of SV40. The EP-half and AT-half origins are indicated. For the EP-half origin, the sequence that is in contact with OBD1, OBD2, and the helicase domain of the LT131 dimer, as revealed in the LT131 dimer-DNA co-crystal structure (PDBid: 4GDF), is indicated. For the AT-half origin, the sequence that is in contact with LT helicase domain, as revealed in the LT helicase hexamer-DNA co-crystal structure, is indicated. (B) LT131 dimer-DNA co-crystal structure, showing how the two LT subunits bind to EP-half origin DNA (PDBid: 4GDF). The EP-half origin sequence is positioned next to the complex structure with the same DNA register to indicate the corresponding DNA sequence bound by the different domains of LT131. With OBD1 binding to PEN1, OBD2 cannot reach PEN2 from subunit 2 because the linker is not long enough. Instead, OBD2 binds to pseudo-PEN1, with an 180° rotation from OBD1. (C) The LT helicase hexamer-DNA co-crystal structure, with the AT-half origin sequences positioned next to the complex structure with the same DNA register to indicate the corresponding DNA sequences bound by LT. For comparison, we tried to best align the register of bound DNA in this panel with that in (B). The PEN1’ and pseudo-PEN’ positions are indicated.
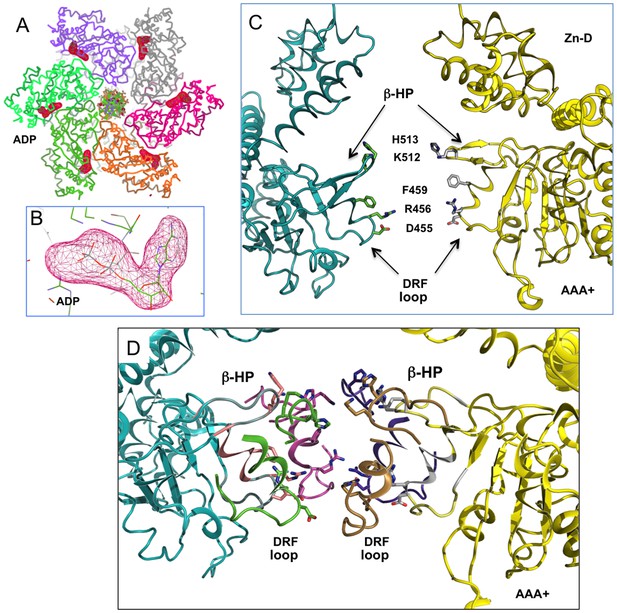
The full occupancy of ADP at all six nucleotide pockets of an LT hexamer and the arrangement of the β-hairpins (β-HP) and DRF loops of the AAA+ domain.
(A) Omit map of ADP density from an LT hexamer-DNA complex structure (one asymmetric unit), showing strong density of the ADP (drawn at 5 sigma level) at all six sites. Even though the electron densities for the six sites are not identical, they are all strong, indicating full occupancy in the DNA-bound hexamer structure, as is the case in all previously determined DNA-free LT hexamer structures. (B) A close-up view at one of the ADP densities, drawn at sigma contour level 5. (C) The β-hairpins (β-HP) and DRF loops of the AAA+ domain in the central channel of a LT hexamer. The β-hairpin residues K512/H513 and the DRF residues D455, R456, F459 are shown as sticks. For clarity, only two opposing subunits of the hexamer are drawn. (D) The near-planar arrangement of the six β-hairpins (β-HP) and DRF loops in the AAA+ domain of the LT hexamer in the DNA complex structure, showing that the β-hairpins and loops are not absolutely planar, but slightly offset. The H513 on the β-hairpin tip is drawn in sticks.
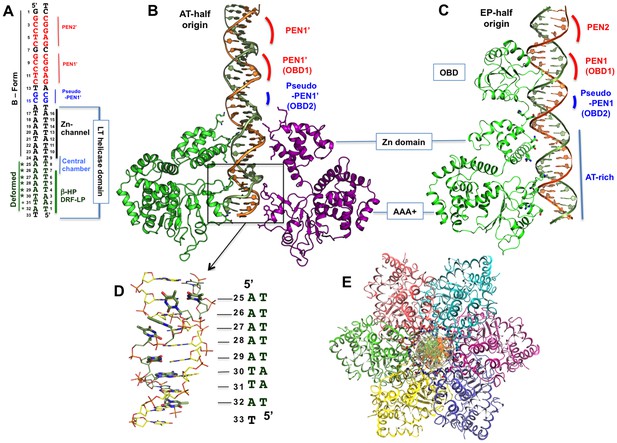
The structure of the LT hexamer helicase in complex with the AT-half origin.
(A) The AT-half origin DNA sequence contains 32 bp with 3’-T overhangs bound by LT hexameric helicase in (B). The side-bars indicate the DNA contacts by different regions within the LT helicase channel, including the Zn-domain, the central chamber, the β-hairpins (β-HP) and DRF loops (DRF-LP). PEN1’ and pseudo-PEN1’ are not occupied by OBD in LT hexamer structure. The deformed region is indicated on the side. (B) The structure of LT131 hexamer bound to the 32-bp AT-half origin DNA. For clarity, only two subunits (one on each side) are shown. The positions of the PEN1’ and pseudo-PEN1’ where OBD1 and OBD2 would bind are indicated. (C) The structure of LT131 dimer bound to the 32-bp EP-half origin DNA (PDBid: 4GDF), showing one of the two subunits using its OBD to bind PEN1. The pseudo-PEN1’ is bound by the OBD of the second subunit of the dimer (Figure 1—figure supplement 1A,B); thehelicase domain of the dimer, including the Zn-domain and the AAA+ domain, binds to the AT-rich region of the EP-half origin. The structures in (B) and (C) are drawn to scale and aligned side-by-side for comparison between the hexamer-DNA and dimer-DNA complexes. (D) The dsDNA segment bound in the AAA+ channel is deformed and locally melted. (E) Top view of the LT hexamer (each subunit in a discrete color) from the N-terminal end, showing dsDNA in the central channel.
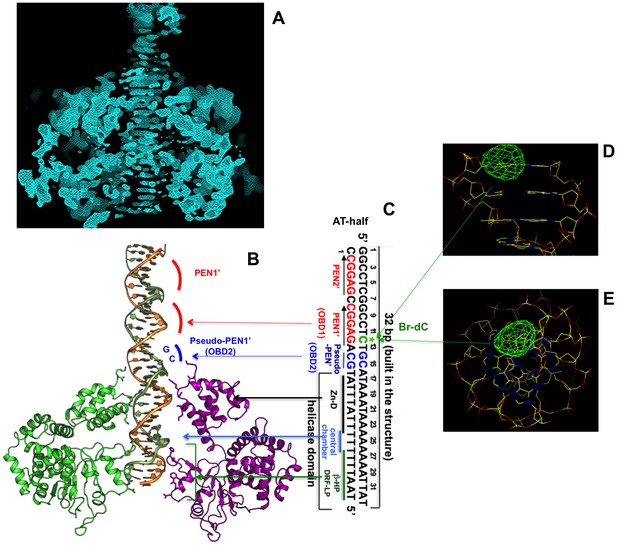
Initial unbiased electron density inside the hexameric channel, and DNA orientation and register confirmed by Bromo-labeled dC (Br-dC) DNA.
(A) A section of the initial electron density map, which was calculated on the basis of the MR LT hexamer model alone prior to any DNA building, showing the dsDNA density with clear nucleotide base steps running through the central channel. The map was calculated as follows (also described in the Materials and methods session). The MR solution model of the LT hexamer was partially deleted on the β-hairpin and DRF loop residues facing the central channel to avoid model bias for the residues that potentially interact with DNA in the channel. The partially deleted MR LT hexamer model was used for calculations of the initial phases and for mask calculation for NCS averaging. Two-domain six-fold NCS averaging was then performed, using a tight mask so that a large central channel space where DNA is located was masked out during NCS averaging, which will avoid averaging the DNA density out in the central channel. The resulting NCS averaged map was drawn here at sigma level 1, which shows the clear DNA base steps in the central channel. (B, C) The AT-half origin DNA inside the hexamer channel (B), and the corresponding DNA sequence (C), with the Bromo-labeled dC (Br-dC at position 12) indicated (colored in green). (D, E) Difference Fourier map of the anomalous data collected from the LT-DNA co-crystals containing the Bromo-labeled dC (Br-dC) DNA at the 12th dC position as shown in (C). The map was drawn with contour sigma level 2.0, showing a strong peak (viewing from two angles) at the expected location for the labeled dC, confirming the DNA orientation and register in the hexamer channel.
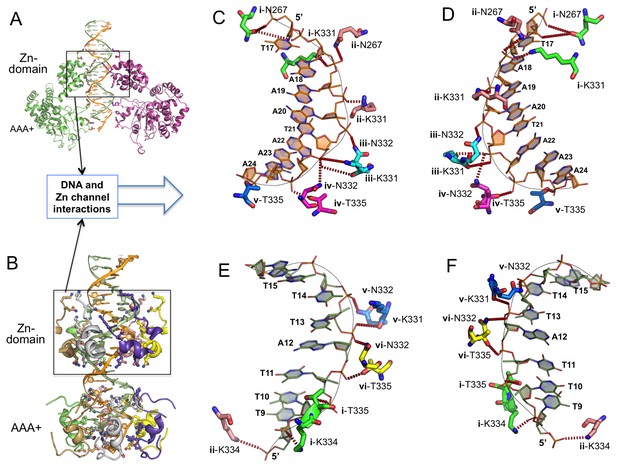
Detailed interactions of origin DNA with the Zn-domain of LT.
(A, B) Two different views of dsDNA inside the LT hexameric channel, one with two complete subunits out of six (A) and the other with the DNA-binding residues on the Zn and AAA+ domains of all six subunits (B). (C, D) Two views tshowing the detailed interactions of the 5’→3’ DNA strand with LT residues inside the Z-domain channel from the N→C direction. The six subunits are labeled as i, ii, iii, iv, v, vi. The residues contacting the DNA are shown as sticks. (E, F) Two views showing the detailed interactions of the 3’→5’ DNA strand with LT residues inside the Z-domain channel from the N→C direction.
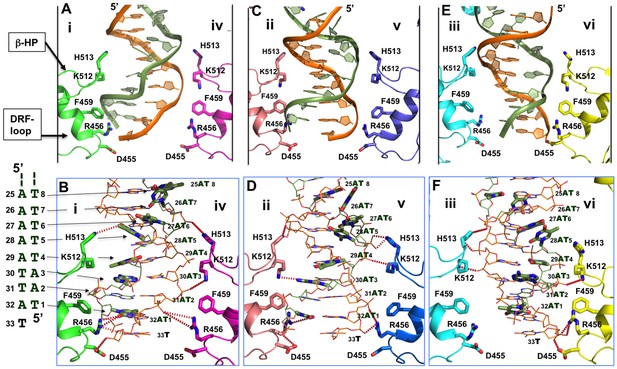
Detailed interactions of origin DNA with the AAA+ domain.
(A, C, E) Detailed interactions of DNA with three pairs of subunits within the LT hexamer: subunit pair i & iv (A), ii & v (C), and iii & vi (E). The β-hairpin residues K512/H513 and DRF loop residues D455/R456/F459 that bind to the DNA are labeled. (B, D, F) The detailed interactions of the oriDNA with the same three pairs of LT subunits shown in (A) [matching B], (C) [matching D], and (E) [matching F]. Dashed lines indicate hydrogen-bond interactions between LT residues and DNA (in the stick model). Some of LT residues are 5–7 Å away from DNA, and would be suitable for the water-mediated formation of hydrogen bonding. In addition, a number of LT residues are within Van der Waals distance of the bound DNA, forming hydrophobic packing interactions; these residues include K512 of subunit i (i-K512), i-F459, iv-F459, ii-F459, v-F459, vi-F459, and vi-H513. The dislocated bases in the disrupted bases-pairs (in the partially melted DNA region) are drawn in thick sticks, with the DNA sequences of this region shown to the left of panel B. The structure shows clearly that identical residues on different subunits make unique contacts with the phosphate backbones and the major or minor gr oves of the DNA; this is possible due to the near-planar arrangement of the six subunits.
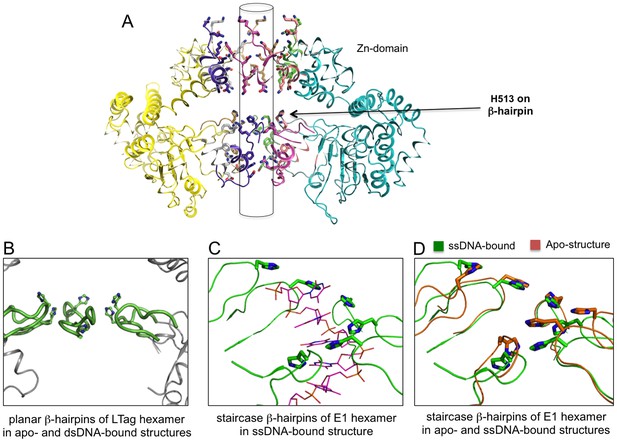
The planar and staircase arrangements of β-hairpins in the hexamer structures of LT and E1.
(A) The near-planar arrangement of the six β-hairpins (indicated by an arrow) in the hexamer channel of LT. Such near-planar arrangement is maintained in all the structures of LT hexamer determined to date, regardless of their dsDNA-bound or DNA-free states. (B) A close-up view of the near-planar arrangement of the six LT β-hairpins from (A), with H513 on the β-hairpin tip shown in sticks. The close-up view reveals that the six H513 and β-hairpins have no real six-fold symmetry. However, a near-planar arrangement is obvious when compared with the staircase arrangement shown in (C). (C) The staircase arrangement of the six β-hairpins (and the tip His residue) in the E1 hexameric channel, showing a six-nucleotide ssDNA (in thin sticks) with each of the six nucleotides havingsimilar interactions with the six β-hairpins (PDBid: 2GXA). (D) Alignment of the β-hairpins in the central channel of E1 hexamer structures in the ssDNA-bound state (PDBid: 2GXA, in green) and the DNA-free state (PDBid: 2V9P, in orange), showing the same staircase arrangement for the β-hairpins, regardless of ssDNA-bound or ssDNA-free state.
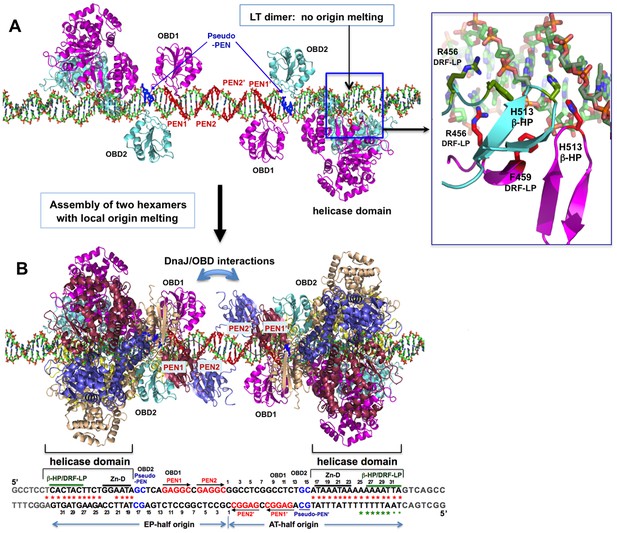
A model for the assembly pathway of the LT double hexamer complex on the viral origin DNA (oriDNA).
(A) The assembly of the intermediate complex containing LT dimer bound to each half of the oriDNA, as predicted on the basis of the co-crystal structure of the LT dimer and EP-oriDNA complex (PDBid: 4GDF). The four PENs (penta-nucleotides) and the pseudo-PENs recognized by OBD are colored in red and blue. In this dimer intermediate, PEN2/PEN2’ near the center are not bound by the OBD from the bound LT dimer, probably because the linker to the helicase domain anchored to the AT-rich origin is too short for the OBD to reach PEN2. Dimerization of LT is not through the ODB, but through the helicase domains. The H513 of the β-hairpin (β-HP) and R456/F459 of the DRF-loop contacting the oriDNA are shown in the boxed inset. The oriDNA is not distorted or meltedin this intermediate complex. (B) Assembly of two head-head hexamers from the dimer intermediate in (A), which forms a near-planar closed-ring instead of the lock-washer open-ring, as predicted based on the hexamer-oriDNA structure here and the prior EM studies (Cuesta et al., 2010; Valle et al., 2006). The origin sequences at the bottom are aligned to the known binding locations on the LT double hexamer, in which the AT-rich sequences of the AT/EP halves are bound by the helicase domain. Red stars indicate the melted/distorted AT-rich sequences as identified biochemically upon assembly of LT double hexamer in the presence of ATP (Borowiec et al., 1990; Borowiec and Hurwitz, 1988). The same studies also showed that the melting of the AT-half origin is much less extensive if ADP was used instead of ATP, even though both ADP and ATP induced similar melting on the EP-half origin. Green stars indicate the melted/distorted sequence in the co-crystal structure of the ADP-bound LT hexamer in complex with the AT-half origin (with the two smaller stars indicating less distortion). It is expected that when the changes in LT conformation are coupled to the binding/hydrolysis of cyclic ATP, the extensively melted/distorted oriDNA will be propagated to form a pair of replication forks and thus to initiate DNA replication.
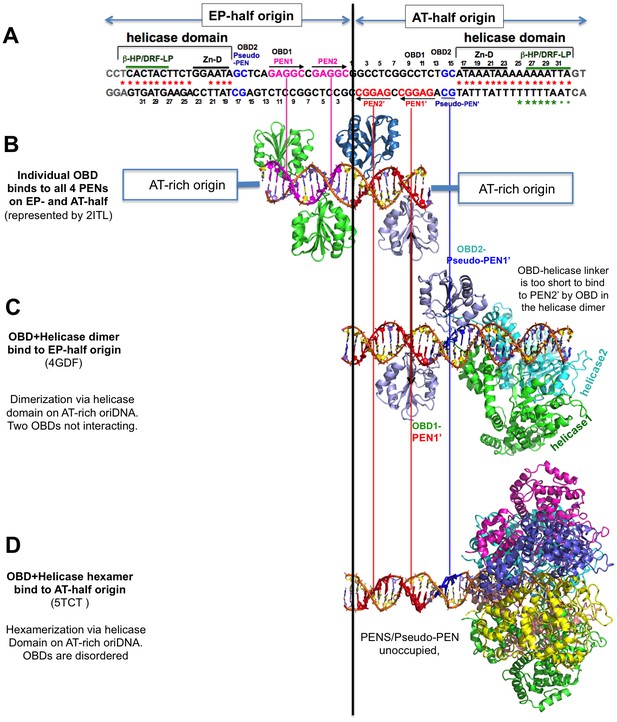
The alignment of the structures of SV40-origin DNA bound to OBD alone (LT131-259) and to the OBD-helicase domain (LT131-627) of LT.
The alignment is based on the origin DNA sequences in the structures with the 64-bp core origin sequence shown at the top. (A) The full core origin sequence of SV40. The EP-half (left) and AT-half (right) origin are capable of supporting hexamer assembly. The four PENs (in red and pink) and the newly identified pseudo-PEN (in blue) are recognized by OBD. The AT-rich sequence interacts with the helicase domain. (B) Structures of the four PENs bound by individual OBDs, (C) the full EP-half origin bound by dimeric LT131-627, and (D) the full AT-half origin bound by hexameric LT131-627. For comparison between the dimeric and hexameric LT131-627, the EP-half origin bound to the dimeric LT131-627 is aligned to the right side with the AT-half. LT131-627 (containing the OBD-helicase domain) is capable of forming a hexamer on each half origin and a double hexamer on a full-origin. Note that when a full hexamer is assembled on each half origin, only the helicase domain hexamerizes, the six OBDs are disordered in the structure, possibly because of the dynamic competition for binding to the three available binding sites (two PENs and one pseudo-PEN). When the LT131-627 dimer is bound to an half origin, only the helicase domain dimerizes, and the two OBD domains that are bound to PEN1 and pseudo-PEN separately, with an 180° inversion, do not interact with each other.
Tables
Crystallographic data and refinement statistics.
| Co-crystal of complex containing LT131, AT-half origin dsDNA, and ADP | |
|---|---|
| Data collection | |
| Space group | P1 |
| Wavelength (λ) | 1.1159 (Å) |
| Cell: a, b, c (Å) | 107.485, 107.587, 107.870 |
| Angles: α, β, γ (°) | 107.133, 106.954, 106.865 |
| Resolution (Å) | 50–2.90 (2.95–2.90) |
| Rmerge (%) | 5.3 (79.7) |
| I / σI | 28.9 (1.52) |
| Completeness (%) | 97.7 (89.1) |
| Redundancy | 3.1 (2.5) |
| Refinement | |
| Resolution (Å) | 50.0–2.9 |
| Unique reflections | 85, 533 |
| Rwork / Rfree (%) | 27.8/30.4 |
| No.atoms | |
| Protein | 17,598 |
| DNA | 1354 |
| Ion | 6 |
| ADP | 162 |
| r.m.s deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.194 |
-
*Highest-resolution shell is shown in parentheses.
The central channel dimensions at the β-hairpins and DRF loops in the hexamer structures of various nucleotide-bound states determined previously and in the DNA-bound state reported here.
| Between β-hairpin tip residues K512 and H513 (Å) | Between DRF loop tip residues D455 and R456 (Å) | |
|---|---|---|
| dsDNA-bound ADP-hexamer (this report) | 19.5, 19.9, 20.1 | 14.8, 14.8, 15.0 |
| dsDNA-free ADP-hexamer (PDB:1SVL) | 11.6, 13.1, 14.6 | 14.0, 16.0, 18.3 |
| dsDNA-free ATP-hexamer (PDB:1SVM) | 6.90, 7.40, 8.90 | 7.90, 8.30, 8.30 |
| dsDNA-free Empty hexamer (PDB:1N25) | 14.5 | 17.9 |
Additional files
-
Supplementary file 1
The detailed interactions between oriDNA and the Zn-domain channel of LT hexamer helicase.
The oriDNA sequences interacting with the Zn-channel of LT are indicated by a black bar on the right panel. The abbreviations and signs in the summary table are as follows:Arrow → donates the interactions between the indicated LT amino acids with the specific DNA residues through the Pi (phosphate via charge or hydrogen bond) from the major groove (MaGv), minor groove (MiGv), or/and directly on the DNA backbones (BkBn).
- https://doi.org/10.7554/eLife.18129.015
-
Supplementary file 2
Detailed interactions between oriDNA and the AAA+ domain channel of LT hexamer helicase.
The oriDNA sequences interacting with the β-hairpin and DRF-loop of LT AAA+ domain are indicated by a green bar on the right panel. The abbreviations and signs in the summary table are as follows: Arrow → donates the interactions between the indicated LT amino acids with the specific DNA residues through the Pi (phosphate via charge or hydrogen bond), bs (base via hydrophobic or hydrogen bond), or bb (backbone via Van der Waals), from the major groove (MaGv), minor groove (MiGv), or/and directly on the DNA backbones (BkBn).
- https://doi.org/10.7554/eLife.18129.016





