Functional dichotomy and distinct nanoscale assemblies of a cell cycle-controlled bipolar zinc-finger regulator
Figures
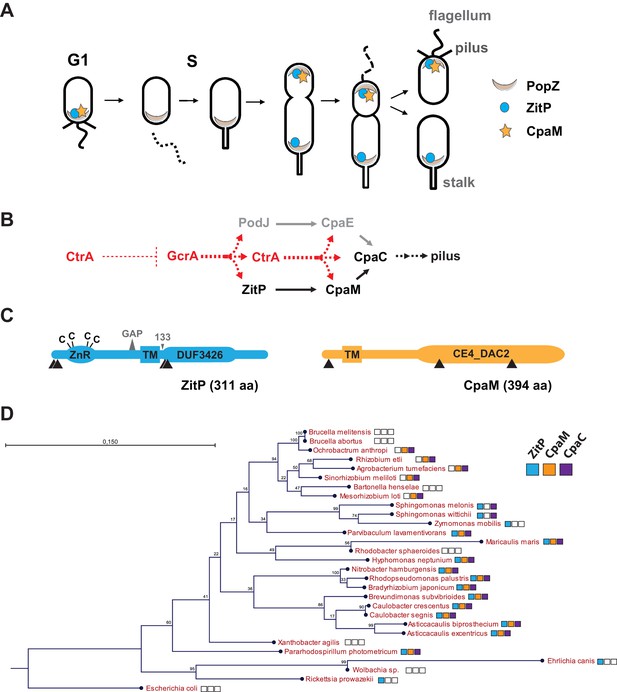
Cell cycle profile and phylogeny of ZitP and CpaM.
(A) Scheme depicting the polarized factors PopZ, ZitP and CpaM during the cell cycle of the dimorphic bacterium C. crescentus. (B) Pilus assembly pathways and global dependencies of the two master cell cycle regulators GcrA and CtrA on the expression of the polar factors PodJ, CpaE, ZitP, CpaM and CpaC that control pilus biogenesis. Red and black dashed lines highlight transcriptional activation and polar recruitment, respectively. (C) Schematic representation (drawn to scale) of ZitP (blue) and CpaM (yellow). ZnR: zinc finger domain; TM: transmembrane domain, C: cysteine. Arrowheads below each protein pinpoint the site of truncation due to transposon insertion in the coding sequence. The large triangle on top of ZitP shows the 2 amino acid residues deleted in the ZitPGAP variant and the small triangle depicts the position of residue 133 where the ZitP coding sequence is truncated in the ZitP1-133 variant. (D) Conservation of ZitP (blue), CpaM (yellow) and CpaC (purple) across the α-proteobacterial clades. The phylogenetic tree was built in CLC Main Workbench (http://www.clcbio.com/products/clc-main-workbench/) from 16S RNA alignments based on the Neighbor Joining method (Juke Cantor substitution model) with 100 bootstrap replicates. Empty boxes mean that no ortholog was found in the genome. Scale bar, 0.15 substitution per site.
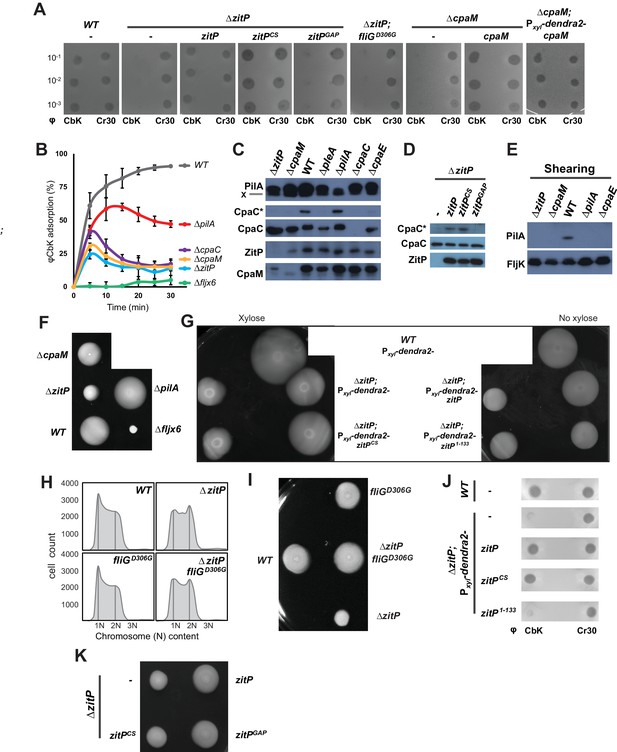
Functional dichotomy in ZitP and effects on polar morphogenesis.
(A) Bacteriophage infection assays of WT, ΔzitP, ΔzitP;fliGD306G and ΔcpaM mutant cells. Cells harbour empty pMT335 or a complementing plasmid (pMT335 backbone) and were grown in the absence of vanillate. No xylose was added to the agar for the phage assay on ΔcpaM; Pxyl-dendra2-cpaM cells. The phages φCbK and φCr30 were spotted with serial dilution on C. crescentus embedded in top agar. Sensitivity to phages is indicated by plaques (lysis). (B) Adsorption kinetics of φCbK to WT and mutant cells. (C) Steady-state levels of ZitP, CpaM, CpaC, modified CpaC (CpaC*) and PilA in WT and mutant cells as determined by immunoblotting. In the PilA immunoblots, the asterisk (*) points to a non-specific band. (D) Immunoblots showing the steady-state levels of monomeric CpaC and CpaC* in ΔzitP cells harbouring pMT335 or derivatives encoding ZitPWT, ZitPCS or ZitPGAP grown in the presence of vanillate (50 µM). (E) Immunoblots showing PilA and FljK abundance in supernatants of WT and various mutant cells. Supernatants were harvested from mid-log cultures after shearing. (F) Swarming motility test performed on soft (0.3%) agar with WT, ΔzitP, ΔcpaM, ΔpilA and Δfljx6 mutant cells. (G) Complementation of the motility defect on swarm (0.3%) agar displayed by the ΔzitP cells expressing Dendra2-ZitP variants from Pxyl at the xylX locus. Xylose was added to the swarm (0.3%) agar as indicated. (H) Flow cytometry of exponential phase WT and ΔzitP cells. N refers to chromosome equivalents. (I) Suppression of the ΔzitP motility phenotype by fliGD306G point mutation as shown on a swarm (0.3%) agar plate. (J) Phage spot tests with φCr30 and φCbK on WT or ΔzitP cells expressing Dendra2-ZitP variants from Pxyl at the xylX locus. Cells were embedded in top agar containing xylose (0.3%). (K) Motility assays of ΔzitP cells expressing WT ZitP (ZitPWT), ZitPCS or ZitPGAP from pMT335. Swarming motility was assessed in absence of vanillate on 0.3% agar.
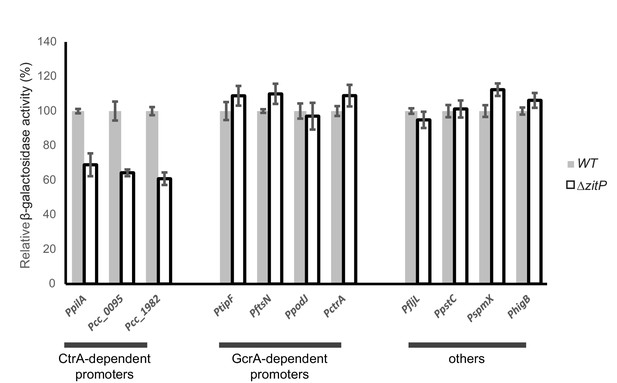
Master regulator-dependent promoters in ΔzitP.
Relative β-galactosidase activity (in percentage) of various lacZ-fused promoters in WT and ΔzitP cells.
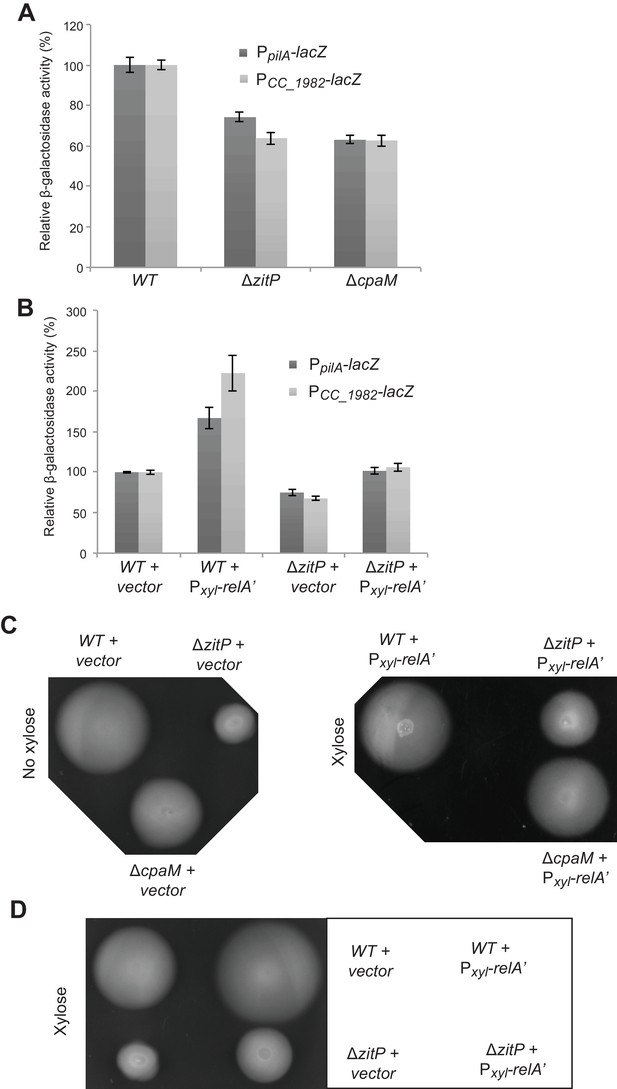
CtrA- and (p)ppGpp-independent influence of the ΔzitP motility defect.
(A) Relative β-galactosidase activity of lacZ-based promoter-probe reporters to the promoters of pilA and CC_1982 in WT, ΔzitP and cpaM cells. (B) Relative β-galactosidase activity of lacZ-based promoter probe reporters to the promoters of pilA and CC_1982 with Pxyl-relA’ (pXTCYC-4-relA′-FLAG) or the pXTCYC-4 control plasmid (vector). (C) Motility test on swarm agar of WT, ΔzitP and ΔcpaM cells transformed with Pxyl-relA’ (pXTCYC-4-relA′-FLAG) or the pXTCYC-4 control plasmid (vector). Xylose was added or not to the agar. (D) Motility test on swarm agar of WT and ΔzitP cells transformed with Pxyl-relA’ (pXTCYC-4-relA′-FLAG) or the pXTCYC-4 control plasmid (vector). Xylose was added to the agar.
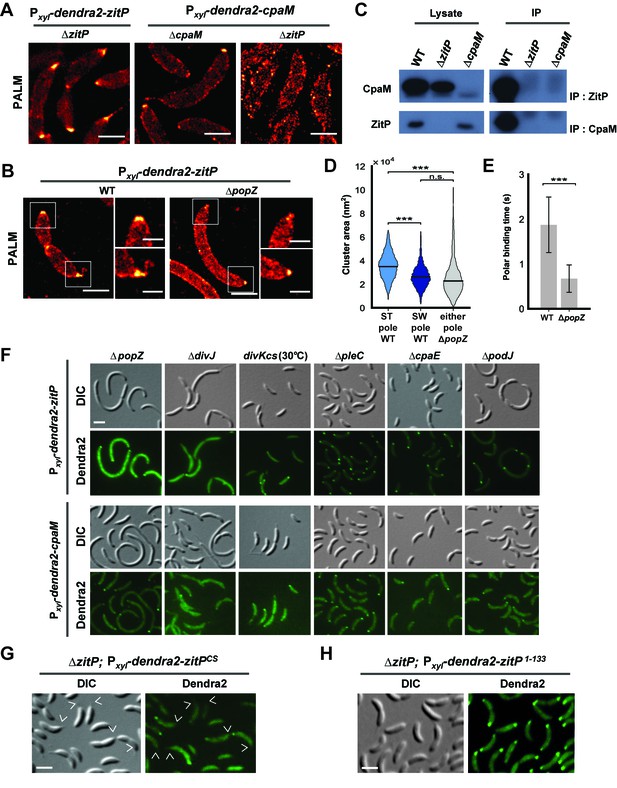
Distinct ZitP nanoscale assemblies and localization determinants.
(A) Photo-activated light microscopy (PALM) imaging of Dendra2-ZitP or Dendra2-CpaM expressed from the xylose-inducible Pxyl promoter on a plasmid integrated at the chromosomal xylX locus in ΔzitP or ΔcpaM cells exposed to xylose 3 hours before imaging. Scale bar: 1 µm. (B) PALM imaging of Dendra2-ZitP in WT or ΔpopZ::Ω cells. We induced expression of Dendra2-ZitP from the xylose-inducible Pxyl promoter on a plasmid integrated at the chromosomal xylX locus by the addition of xylose 3 hours before imaging. Scale bar: 1 µm. Scale bar of zoomed images: 0.5 µm. (C) Co-immunoprecipitation (co-IP) of ZitP or CpaM with polyclonal antibodies to CpaM or ZitP, respectively. Immunoprecipitates and cell lysates from WT, ΔzitP or ΔcpaM cells were probed for the presence of ZitP or CpaM. (D) Projected area of the Dendra2-ZitP polar complex as determined by PALM from Dendra2-ZitP expressed in WT and ΔpopZ::Ω cells. Black lines indicate medians. Statistical significance from Mood’s median test: n.s, p>0.05; ***p<0.001. (E) ZitP polar binding times in WT and ΔpopZ::Ω cells, measured via single particle tracking PALM. Error bars indicate 95% confidence interval of the fit to the data (Figure 3—figure supplement 6D). Statistical significance from a 2 sample t-test: ***p=p<0.001. (F) Epifluorescence (Dendra2) and Nomarski (DIC) images depicting the localization of Dendra2-ZitP or Dendra2-CpaM in ΔpopZ::Ω, ΔdivJ, divKcs, ΔpleC, ΔcpaE or ΔpodJ cells. Expression of Dendra2-ZitP or Dendra2-CpaM was induced from the chromosomal xylX locus with xylose 4 hours before imaging. Scale bars: 1 µm. (G) (H) Epifluorescence (Dendra2) and Nomarski (DIC) images depicting the localization of the motility-deficient and pilus-proficient Dendra2-ZitPCS variant (G) or the motility-proficient and pilus-deficient Dendra2-ZitP1-133 variant (H) in ΔzitP cells. Arrow heads pinpoint stalked poles. We induced expression of Dendra2-fusions from the xylose-inducible Pxyl promoter on a plasmid integrated at the chromosomal xylX locus by the addition of xylose 4 hours before imaging. Scale bars: 1 µm.
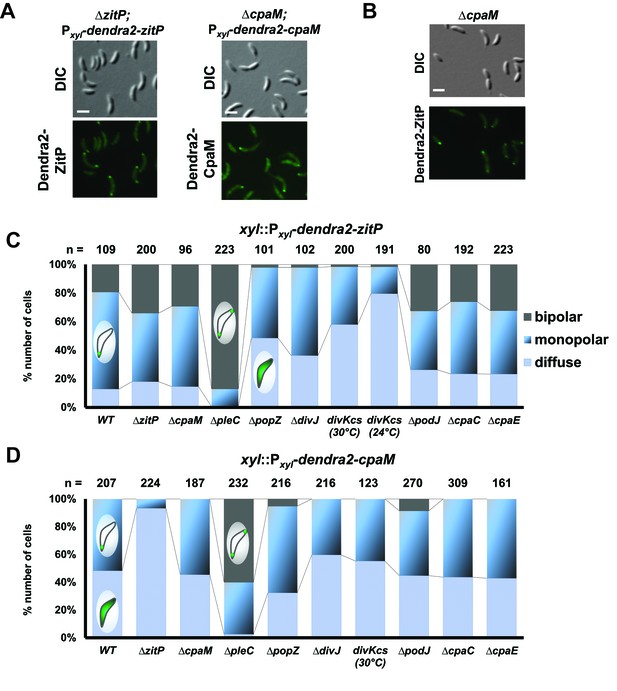
Extrinsic determinant for the localization of ZitP and CpaM.
(A) Epifluorescence (Dendra2) and Nomarski (DIC) images depicting the localization of Dendra2-ZitP and Dendra2-CpaM variants in asynchronious ΔzitP or ΔcpaM cells, respectively. We induced expression of Dendra2 fusions expressed from the xylose-inducible Pxyl promoter on a plasmid integrated at the chromosomal xylX locus. Scale bars: 1 µm. (B) Subcellular localisation of Dendra2-ZitP in the ΔcpaM mutant. Cells were imaged in epifluorescence (GFP channel) and bright field mode (DIC). We induced expression of Dendra2-ZitP from Pxyl on plasmids integrated at the chromosomal xylX locus by the addition of xylose 4 hours before imaging. Scale bar: 1 µm. (C) (D) Quantification of Dendra2-ZitP (C) or Dendra2-CpaM (D) localization states (diffuse, monopolar or bipolar) in WT or polarity mutants cells. Dendra2 fusions were expressed from the xylose-inducible Pxyl promoter on a plasmid integrated at the chromosomal xylX locus. The total cell count (n) for each strain is shown above related stacked bars. Values are expressed in percentage of whole cell population.
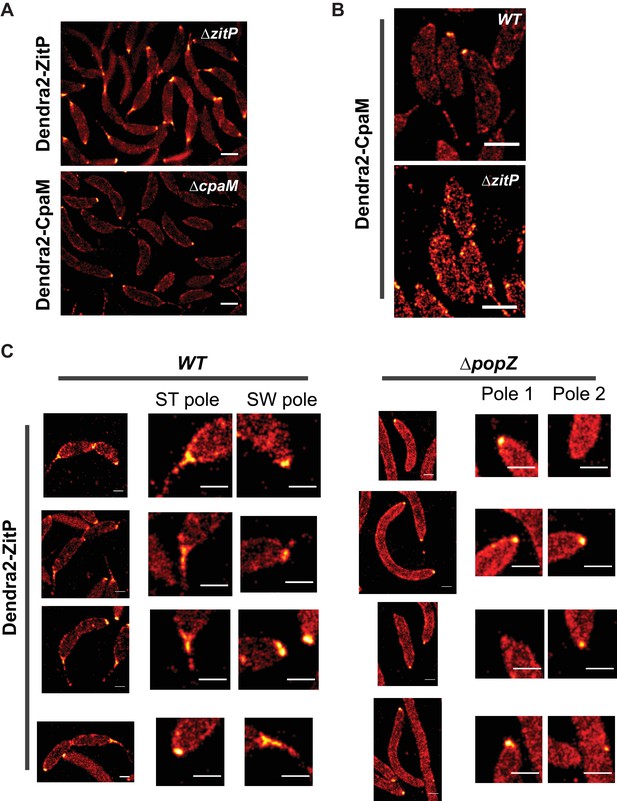
ZitP and CpaM polar localization by PALM.
(A) PALM images of ZitP or CpaM localization in ΔzitP and ΔcpaM cells, respectively. We induced expression of Dendra2-fusions from the xylose-inducible Pxyl promoter on a plasmid integrated at the chromosomal xylX locus by the addition of xylose 3 hours before imaging. Scale bars: 0.5 µm. (B) PALM images of CpaM localization in WT and ΔzitP cells. We induced expression of Dendra2-fusions from the xylose-inducible Pxyl promoter on a plasmid integrated at the chromosomal xylX locus by the addition of xylose 3 hours before imaging. Scale bars: 1 µm. (C) ZitP localization, with zoomed images of poles in WT and ΔpopZ::Ω cells. Scale bar 0.5 µm.
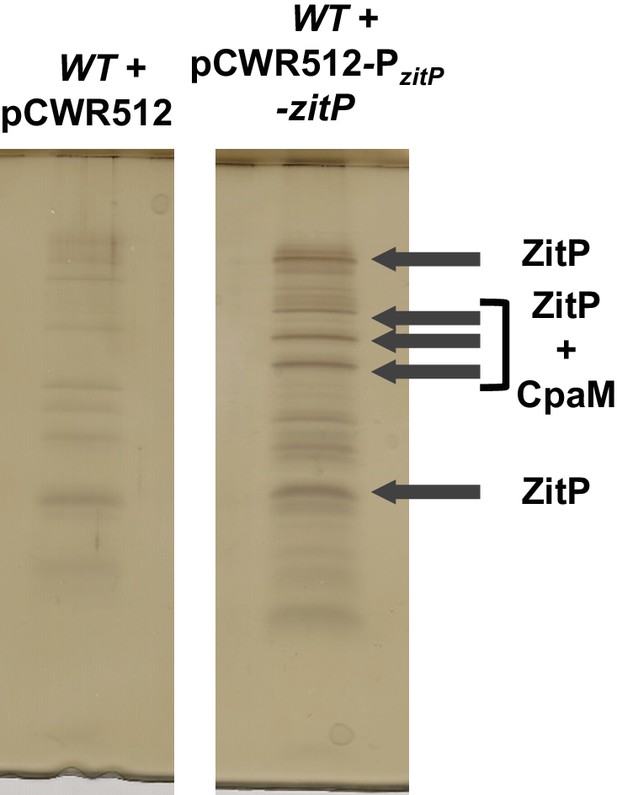
Tandem affinity purification of ZitP.
Tandem affinity purification (TAP) performed on WT cell extracts bearing an empty pCWR512 plasmid (control) or the Pvan-zitP-TAP plasmid. After electrophoresis of TAP extracts, the gel was silver-stained as guided by the manufacturer (SilverQuest, Invitrogen). Arrows indicate bands that were extracted and sent for mass spectrometry analyses. They indicate as well the main identified proteins.
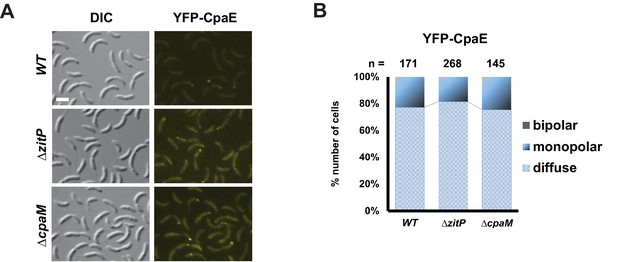
CpaE localization in ΔzitP and ΔcpaM mutant cells.
(A) Epifluorescence (YFP-CpaE) and Nomarski (DIC) images depicting the localization of the pilus component CpaE N-terminally fused to YFP expressed form the native cpaE locus (yfp-cpaE) in WT, ΔzitP and ΔcpaM cells. The scale bars represent 1 µm. (B) Quantification of YFP-CpaE localization profile (diffuse, monopolar or bipolar) in the aforementioned strains. The total cell count (n) for each strain is shown above related stacked bars. Values are expressed in percentage of whole cell population.
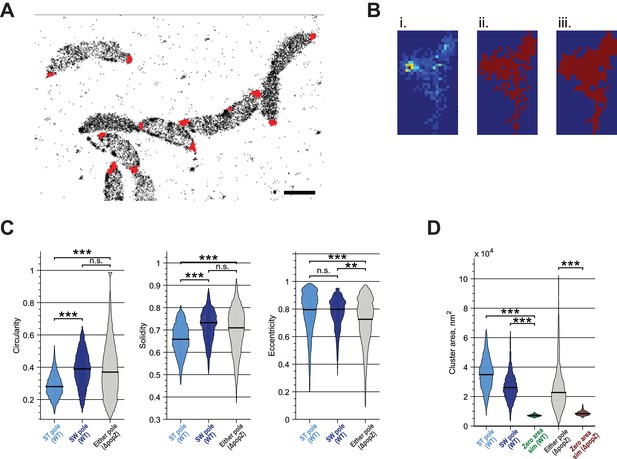
Quantitative analysis of ZitP cluster shape and area.
(A) Measurement of polar cluster area. Automated clustering of Dendra2-ZitP localization using DBSCAN. Red dots, identified clusters; black crosses, low-density localizations outside of polar clusters. (B) Image processing operations for area measurement. Identified clusters (i) were converted to a binary image (ii) which was then processed via morphological closing (iii) to make the cluster area measurement less sensitive to noise and molecule sampling rate. (C) Measured circularity, solidity and eccentricity of WT stalked pole, WT other (swarmer) pole and polar foci in ΔpopZ::Ω cells. (D) Measured area of WT stalked pole, WT other (swarmer) pole and polar foci in ΔpopZ::Ω cells compared to the observed area of simulated zero-area clusters. Observed zero-area cluster size is non-zero due to experimental noise. Stars indicate statistical significance: n.s, p>0.05; *p<0.05; **p<0.01; ***p<0.001.
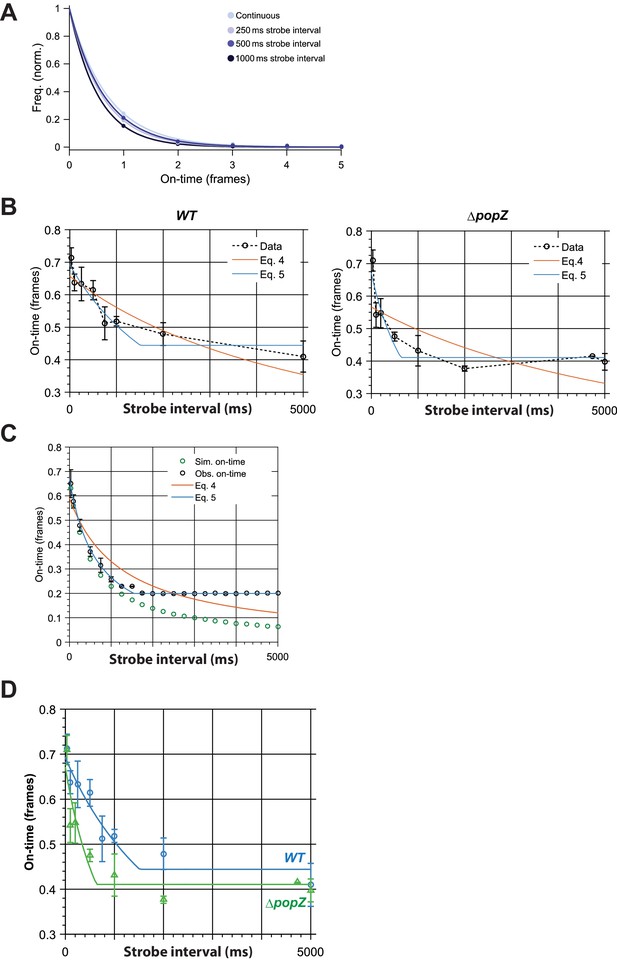
Binding time estimation by stroboscopic single particle tracking of ZitP.
(A) Exemplar histograms of effective on-time (ie. combination of actual binding lifetime with photobleaching lifetime) in different time-lapse conditions for Dendra2-ZitP in WT cells. Lines are fits by a single exponential model. (B) Observed effective on-time of polar Dendra2-ZitP as a function of time-lapse duration in WT and ΔpopZ cells. Fitting of the data with Eq 4 (Gebhardt model) shows large systematic errors. By accounting for finite camera integration time (Eq 5) we obtained good fits to the data. Error bars indicate 1 s.d. (C) Simulation showing the effect of finite camera integration time on observed on-time. Observed on-time (obs. on-time) shows a strong plateau at a minimum observable on-time, compared to the true on-time (sim. on-time), confirming that the plateau observed experimentally most likely arises from finite camera integration time. By accounting for this effect (Eq 5) we obtained good fits to the data. Error bars indicate 1 s.d. (D Comparison of observed effective on-time of polar Dendra2-ZitP as a function of time-lapse duration in WT and ΔpopZ cells, showing fitting with the revised binding time model (Eq 5). Error bars indicate 1 s.d.
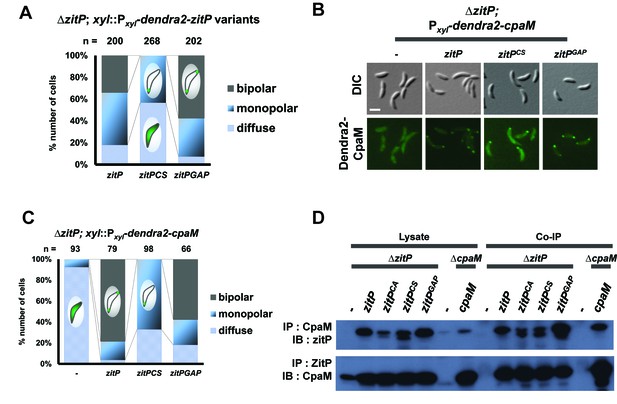
Intrinsic determinants for ZitP localization and function.
(A) Epifluorescence (Dendra2) and Nomarski (DIC) images depicting the localization profile of Dendra2-CpaM in ΔzitP cells complemented with ZitP-expression plasmids. ZitPWT, ZitPCS or ZitPGAP were expressed from pMT335 (without vanillate). We used empty pMT335 (-) as a control. Scale bars: 1 µm. (B) Quantification of Dendra2-CpaM localization states (diffuse, monopolar or bipolar) in ΔzitP cells expressing ZitPWT, ZitPCS or ZitPGAP from pMT335 (without vanillate). We used empty pMT335 (-) as a control. Dendra2 fusions were expressed from the xylose-inducible Pxyl promoter on a plasmid integrated at the chromosomal xylX locus. The total cell count (n) for each strains are shown above related stacked bars. Values are expressed in percentage of whole cell population. (C) Reciprocal co-immunoprecipitation (IP) of ZitP variants or CpaM protein with polyclonal antibodies to CpaM or ZitP, respectively. Immunoprecipitates and cell lysates were probed for the presence of ZitP or CpaM by immunoblotting (IB). Extracts were made from ΔzitP and ΔcpaM cells expressing ZitPWT, ZitPCS, ZitPGAP or CpaM from pMT335 grown without vanillate. (D) Quantification of Dendra2-ZitPCS and Dendra2-ZitPGAP localization states (diffuse, monopolar or bipolar) in the ΔzitP cells from the xylose-inducible Pxyl promoter on a plasmid integrated at the chromosomal xylX locus. The total cell count (n) is shown above the stacked bar. Value is expressed in percentage of whole cell population.
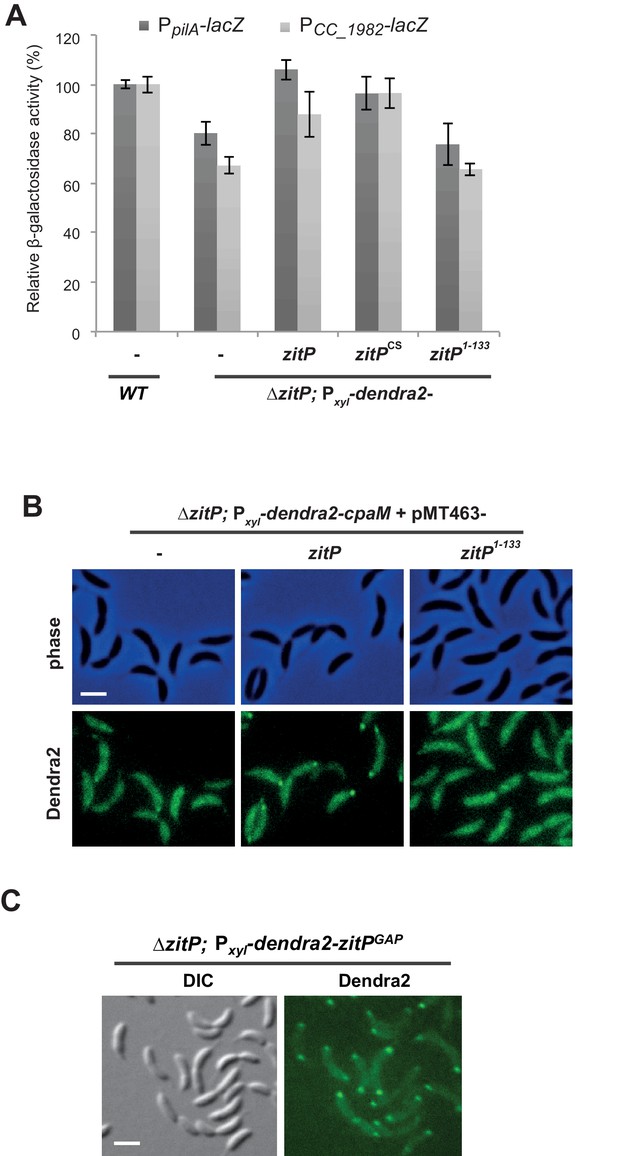
Effect of DUF3426 on ZitP function.
(A) Relative β-galactosidase activity of lacZ-based promoter-probe reporters to the pilA and CC_1982 promoter in WT and zitP cells expressing Dendra2-ZitP variants from Pxyl at the xylX locus. Xylose was added to the medium. (B) Epifluorescence (Dendra2) and phase images depicting the localization profile of Dendra2-CpaM in ΔzitP cells complemented with pMT463-derived plasmids expressing either WT ZitP (ZitPWT) or ZitP1-133. We used empty pMT463 (-) as a control. Scale bars: 1 µm. (C) Epifluorescence (Dendra2) and phase contrast overlays depicting the localization of Dendra2- ZitPGAP from the xylX locus in ΔzitP cells. Scale bars: 1 µm
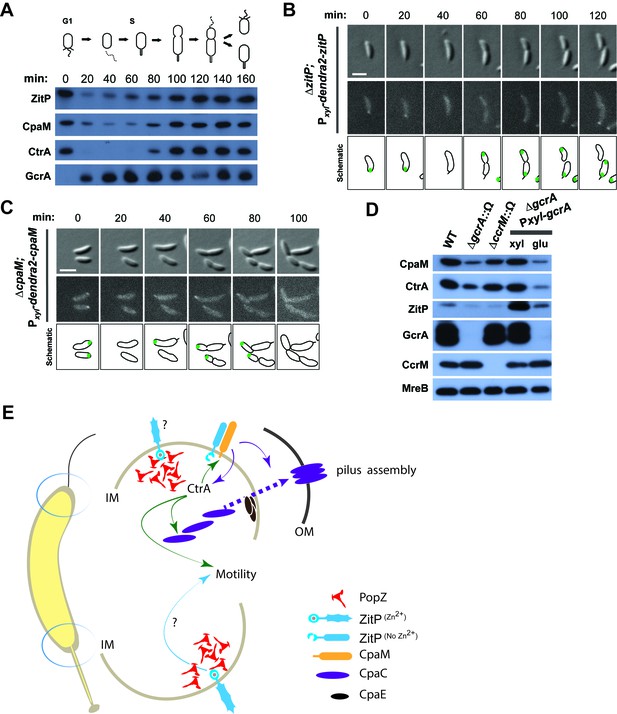
Cell cycle regulation of ZitP and CpaM localization.
(A) Immunoblots showing the levels of ZitP, CpaM and master cell cycle regulators along the C. crescentus cell cycle in a synchronized WT population. The upper scheme depicts C. crescentus cell cycle stages. (B) (C) Epifluorescence (Dendra2) and Nomarski (DIC) images depicting the localization of Dendra2-ZitP (B) and Dendra2-CpaM (C) in synchronized ΔzitP or ΔcpaM cells, respectively. We induced expression of Dendra2 fusions expressed from the xylose-inducible Pxyl promoter on a plasmid integrated at the chromosomal xylX locus. Schematic drawings highlight Dendra2 localizations. After synchronization, cells were resuspended in M2G and imaged every 20 minutes. Scale bars: 1 µm. (D) Steady-state levels of ZitP, CpaM, CtrA, GcrA, CcrM and MreB (control) in WT, gcrA and ccrM mutant cells. Xylose (0.3%, xyl) or glucose (0.2%, glu) were supplemented to the medium in order to induce/deplete GcrA in ΔgcrA xylX::Pxyl-gcrA cells. (E) Schematic representation of the two Caulobacter cell poles. At the stalked pole, the PopZ matrix promotes the recruitment of ZitP. The Zn2+-bound zinc-finger domain of ZitP prevents ZitP/CpaM association and influences CtrA activity and swarming motility. At the opposite pole, the inactive Zn2+-unbound zinc-finger domain allows the formation of the ZitP/CpaM complex and the export and assemblage of CpaC in the outer membrane (OM)independently of PopZ.
Additional files
-
Supplementary file 1
Bacterial strains, plasmids and oligonucleotides list
- https://doi.org/10.7554/eLife.18647.017





