Ubiquitin-dependent folding of the Wnt signaling coreceptor LRP6
Figures
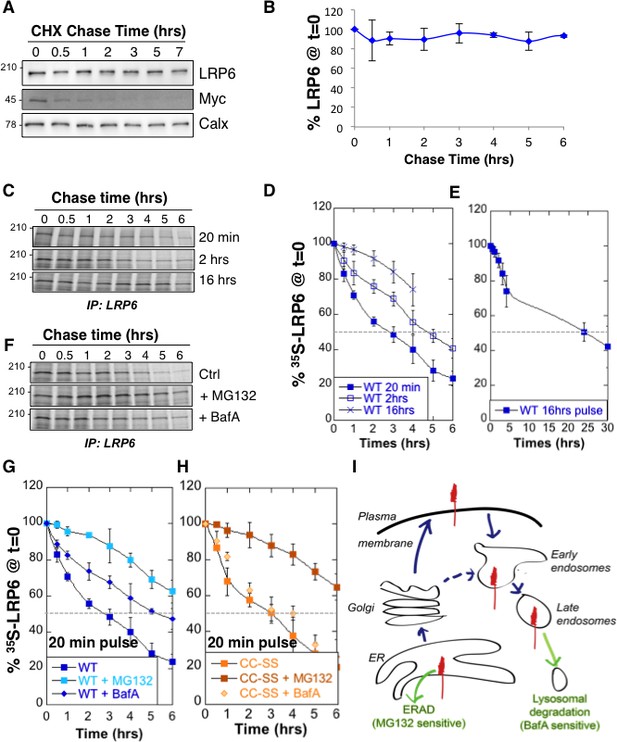
LRP6 undergoes rapid degradation following synthesis in the ER but is stable once mature.
(A) representative western blot of a cycloheximide (CHX) chase in RPE1 cells. 40 µg of total cell extracts from RPE1 cells were loaded per lane, analyzed by SDS-PAGE followed by Western blotting against endogenous LRP6, calnexin (Calx), a stable protein and Myc, a short lived protein. (B) Experiments as in A were quantified by ImageJ software, n = 3. CDE: RPE1 cells were submitted to metabolic 35S Cys/Met labeling for different times and subsequently chased for different times. Endogenous LRP6 was immunoprecipitated with an anti-LRP6 antibody. A representative experiment in shown in (C). Autoradiograms were quantified using the Typhoon imager and means of different experiments were calculated (D and E). Error bars represent standard deviation (n = 6 for the 20 min pulse; n = 4 for the 2 hr and 16 hr pulses). FG: RPE1 cells were treated or not with MG132 or Bafilomycin A and subsequently submitted, in the presence or not of the drugs, to metabolic a 20 min 35S-Cys/Met pulse followed by different chase times. A representative experiment in shown in (F). Errors represent standard deviation (n = 4 for MG132; n = 3 for Bafilomycin A, BafA, the WT control curve corresponds to that shown in Figure 1D). (G) Hela cells transiently expressing myc-tagged palmitoylation deficient LRP6 (CC–SS) were submitted to metabolic 35S Cys/Met labeling for different times and subsequently chased for different times. LRP6 was subsequently immunoprecipitated using an anti-myc antibody. Errors represent standard deviation (n = 3). H: Cartoon depicting the two major cellular degradation pathways for membrane proteins: ERAD (blocked by MG132) and lysosomal pathway (blocked by Bafilomycin A).
-
Figure 1—source data 1
Numeric data for graphs of Figure 1B,D,E and F and 1 hr.
- https://doi.org/10.7554/eLife.19083.004
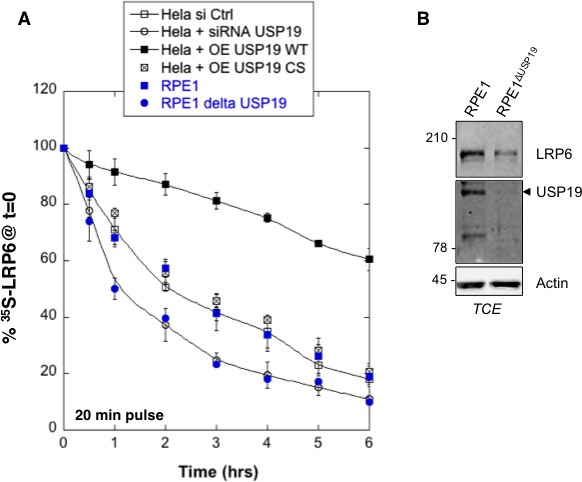
Variation in USP19 cellular amount influences LRP6 degradation rates.
(A) Metabolic 35SCys/Met pulse chase experiment (20 min pulse) in the following conditions: immunoprecipitation of myc-LPR6 in HeLa cells transiently expressing myc-LRP6 wild type (WT) upon control silencing (HeLa si Ctrl, n = 7) vs. usp19 silencing (HeLa + siRNA USP19, n = 6) or upon co-overexpression of myc-LRP6 wild type and wild type GFP-tagged USP19 (HeLa + OE USP19 WT, n = 6) vs. catalytically inactive GFP-tagged USP19 (HeLa + O.E. USP19 CS, n = 6); immunoprecipitation of endogenous LRP6 in RPE1 cells wild type (RPE1, n = 4) vs. RPE1 knout-out cells for usp19 gene (RPE1 delta USP19, n = 3). (B) Total cell extract of wild type RPE1 and RPE1 knout-out cells for usp19 gene revealed with USP19, LRP6 and Actin antibodies.
-
Figure 1—figure supplement 1—source data 1
Numeric data for graphs of Figure 1—figure supplement 1A.
- https://doi.org/10.7554/eLife.19083.006
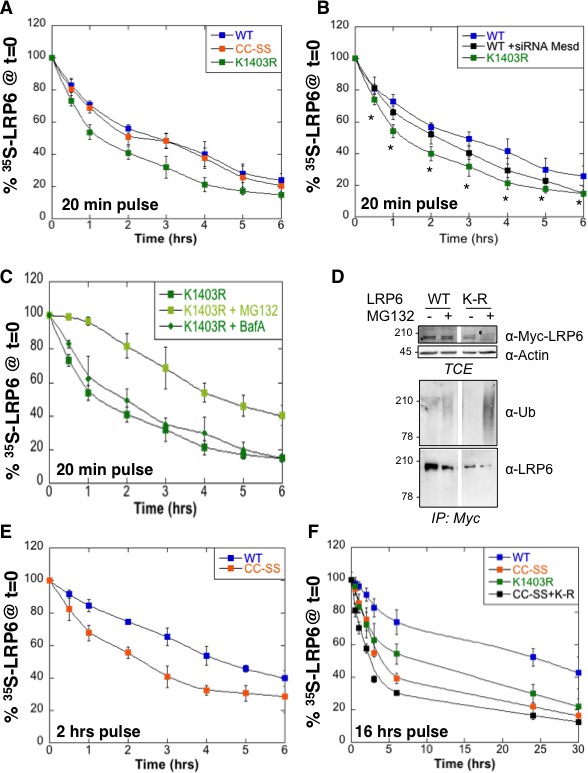
Mutation of the palmitoylation sites and the Lys-1403 ubiquitination site accelerate LRP6 targeting to ERAD.
(A) Metabolic 35SCys/Met pulse chase experiment (20 min pulse) on transiently expressed myc-LRP6 wild type (WT, curve corresponding to the one in Figure 1D), palmitoylation deficient (CC-SS, n = 3) or K1403R (KR, n = 6) mutants in HeLa cells. (B) Metabolic 35SCys/Met pulse chase experiment (20 min pulse) on transiently expressed myc-LRP6 wild type (WT, n = 7), or K1403R (K1403R, n = 7) mutant in HeLa cells silenced or not for mesd gene (siRNA mesd, n = 3). Errors represent standard deviation,*<p=0.05 calculated between LRP6 WT and K1403R. (C) Metabolic 35SCys/Met pulse chase experiment (20 min pulse) on transiently expressed myc-LRP6K1403R in HeLa cells supplemented or not (n = 6, curve corresponding to the one in Figure 2A) with MG132 (K1403 + MG123, n = 3) or Bafilomycin A (K1403 + BafA, n = 3). (D) Immunoprecipitation of myc-tagged LRP6 Wild Type (WT) and K1403 mutant (KR) revealed with anti-Ubiquitin antibody, with or without MG132 treatment. (E) Metabolic 35SCys/Met pulse chase experiment (2 hr pulse) on transiently expressed myc-LRP6 wild type (WT, n = 4, curve corresponding to the one in Figure 1D) or palmitoylation deficient (CC-SS, n = 3) in HeLa cells. (F) Metabolic 35SCys/Met pulse chase experiment (16 hr pulse) on transiently expressed myc-LRP6 wild type (WT, curve corresponding to the one in Figure 1D), palmitoylation deficient (CC-SS, n = 3), K1403R (K1403R, n = 3) or K1403R in the palmitoyl deficient background (CC-SS + KR, n = 3) mutants in HeLa cells.
-
Figure 2—source data 1
Numeric data for graphs of Figure 2A,B,C and E,F.
- https://doi.org/10.7554/eLife.19083.008
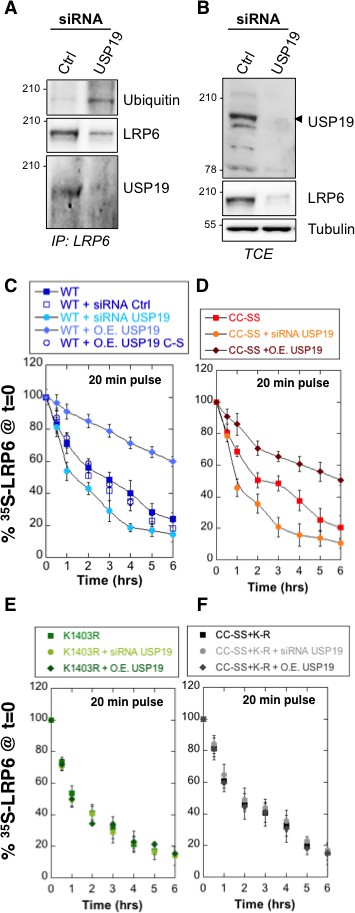
De-ubiquitination of LRP6 by USP19 on Lys-1403 promotes LRP6 biogenesis.
(A) Immunoprecipitation of endogenous LRP6 and (B) cellular level in RPE1 cells upon usp19 silencing. C/D/E/F: Metabolic 35SCys/Met pulse chase experiment (20 min pulse) on transiently expressed myc-LRP6 wild type (WT), palmitoylation deficient mutant (CC–SS), K1403R mutant (K1403R) or K1403R mutation in the palmitoyl deficient background (CC-SS + KR) mutants in HeLa cells upon over expression of GFP-tagged USP19 (O.E. USP19) or GFP-tagged USP19 catalytically inactive (O.E. USP19 C-S) or upon usp19 gene silencing (siRNA USP19). (C) WT n = 6, same as in Figure 1D, other conditions n = 3, (D) (CC-SS curve corresponding to the one in Figure 2A) and (F) all conditions n = 3, (E) K1403R n = 6, other conditions n = 3.
-
Figure 3—source data 1
Numeric data for graphs of Figure 3C, D and E, F.
- https://doi.org/10.7554/eLife.19083.010
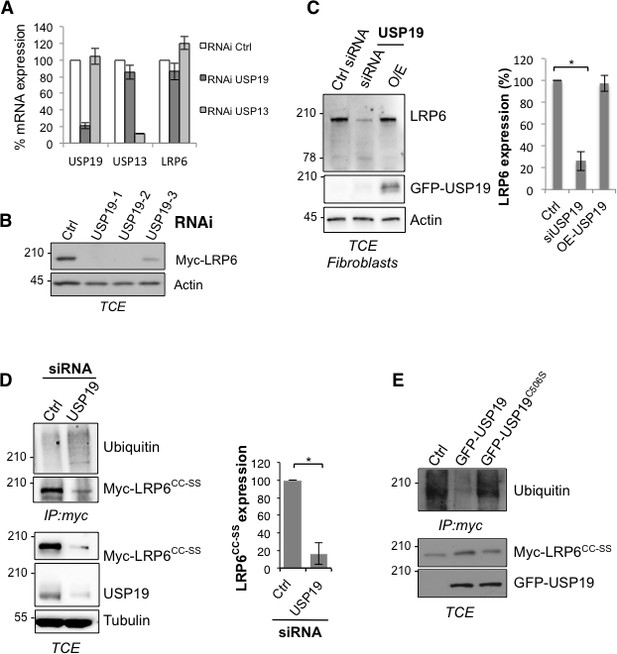
Variation in USP19 cellular amount influences LRP6 ubiquitination state.
(A) RT-PCR detecting usp19, usp13 and LRP6 mRNA expression level upon silencing of usp19 and usp13 genes with specific RNAi. Errors represent standard deviation. B Total cell extracts of Hela transiently expressing Myc-LRP6 and silenced with 3 different RNAi targeting usp19 gene. C) Total cell extract of primary fibroblasts silenced for usp19 gene or overexpressing GFP-tagged USP19 revealed with GFP, LRP6 and Actin antibodies. Quantification of LRP6 cellular amount is shown on the right. Errors represent standard deviation (n = 3) and *<p=0.05. (C) Immunoprecipitation of transiently expressed palmitoylation deficient myc-LRP6 (Myc-LRP6CC-SS) upon usp19 silencing. Quantification of Myc-LRP6CC-SS cellular amount in TCE is shown on the right. Errors represent standard deviation (n = 3) and *<p=0.05. (D Immunoprecipitation of transiently expressed palmitoylation deficient myc-LRP6 (Myc-LRP6CC-SS) upon overexpression of GFP-tagged USP19 vs. GFP-tagged USP19 (GFP-USP19) catalytically inactive mutant (GFP-USP19C506S).
-
Figure 3—figure supplement 1–source data 1
Numeric data for graphs of Figure 3—figure supplement 1A, C .
- https://doi.org/10.7554/eLife.19083.012
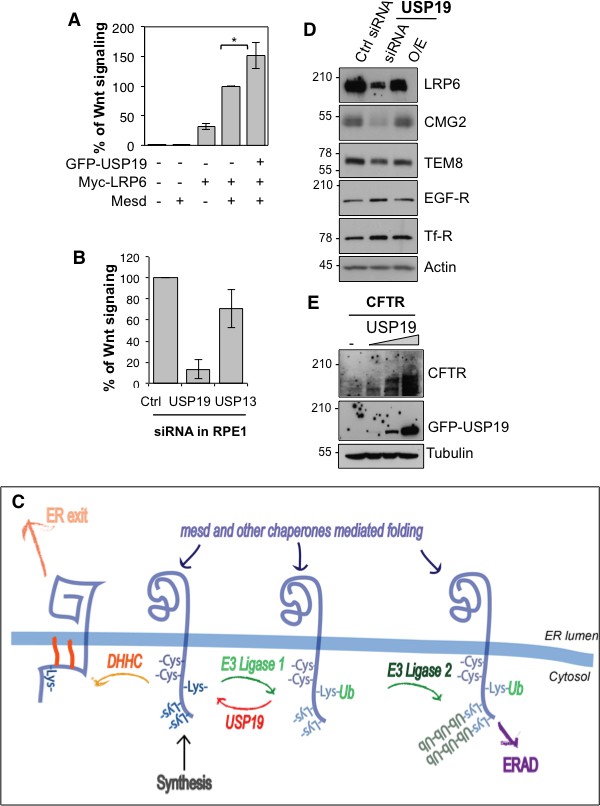
USP19 controls the Wnt signaling capacity of the cell.
(A) Wnt signaling measured in HEK293 cells carrying the TOPFLASH reporter assay, transiently transfected for the indicated constructs (n = 5) (B) Wnt signaling measured in RPE1 cells stably expressing lentiviral vector possessing a 7xTCF-FFluc upon usp19 or usp13 silencing (n = 4). (C) Working model (described in the text). (D) cellular level of the indicated endogenous proteins in RPE1 cells upon usp19 silencing (siRNA) or overexpression (O.E.) of GFP-tagged USP19. (E) cellular level of transiently co-transfected CFTR (constant amount) and GFP-tagged USP19 (increasing amount) in RPE1 cells.
-
Figure 4—source data 1
Numeric data for graphs of Figure 4A, B.
- https://doi.org/10.7554/eLife.19083.014
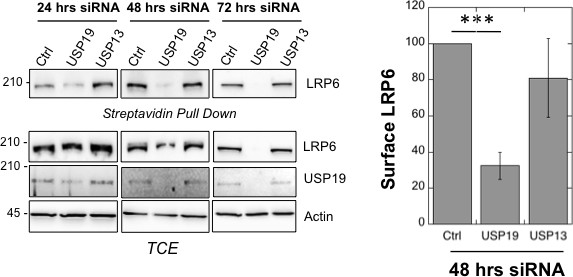
usp19 silencing leads to decrease in LRP6 cell surface expression.
Surface Biotinylation assay performed in RPE1 cells upon 24, 48 and 72 hr of usp19 or usp13 gene silencing. Quantification of endogenous LRP6 surface expression at 48 hr of gene silencing in Streptavidin-mediated pull down is shown above the western blot. Errors represent standard deviation (n = 3) and ***<p 0.0005.
-
Figure 4—figure supplement 1–source data 1
Numeric data for graphs of Figure 4—figure supplement 1.
- https://doi.org/10.7554/eLife.19083.016





