Distinct roles for extracellular and intracellular domains in neuroligin function at inhibitory synapses
Figures
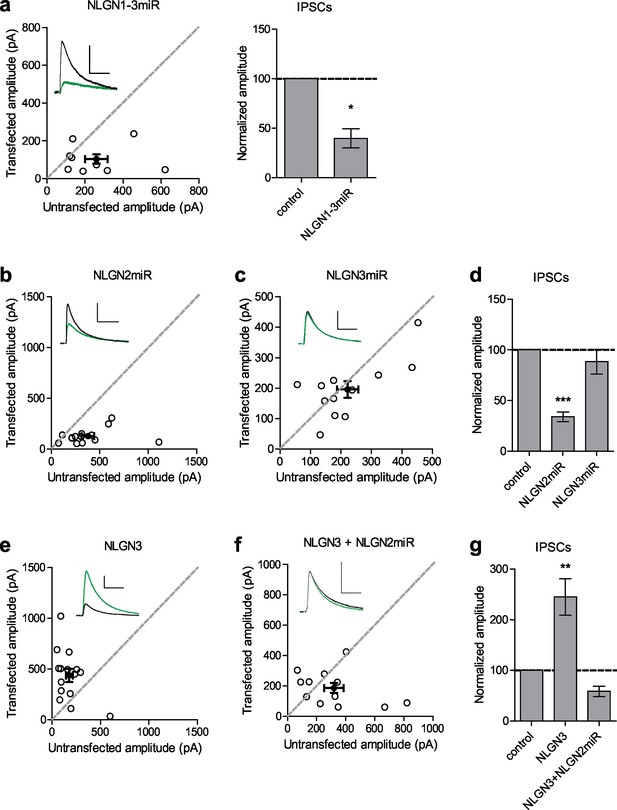
Neuroligin 2 is the critical neuroligin at inhibitory synapses.
(a) Knockdown of endogenous neuroligins 1–3 reduces inhibitory responses compared to untransfected control cells (*p=0.0391, n = 9). (b) Scatter plot showing reduction in IPSCs in NLGN2 microRNA-transfected neurons compared to untransfected controls (***p=0.0002, n = 15). (c) Scatter plot showing no reduction in IPSCs in NLGN3 microRNA-transfected neurons compared to untransfected controls (p=0.3013, n = 12). (d) Summary graph of b and c. (e) Scatter plot showing overexpression of NLGN3 enhances IPSCs compared to untransfected controls (**p=0.0084, n = 15). (f) Scatter plot showing overexpression of NLGN3 combined with NLGN2 knockdown fails to enhance IPSCs (p=0.2661, n = 12). (g) Summary graph of e and f. For panels a-c and e-f, open circles are individual pairs, filled circle is mean ± s.e.m. Black sample traces are control, green are transfected. Scale bars represent 100 pA and 50 ms. For panels a, d, and g summary graph plots mean transfected amplitude ± s.e.m, expressed as a percentage of control amplitude. Significance above each column represents pairwise comparison between transfected and untransfected cells. See also Figure 1—figure supplement 1.
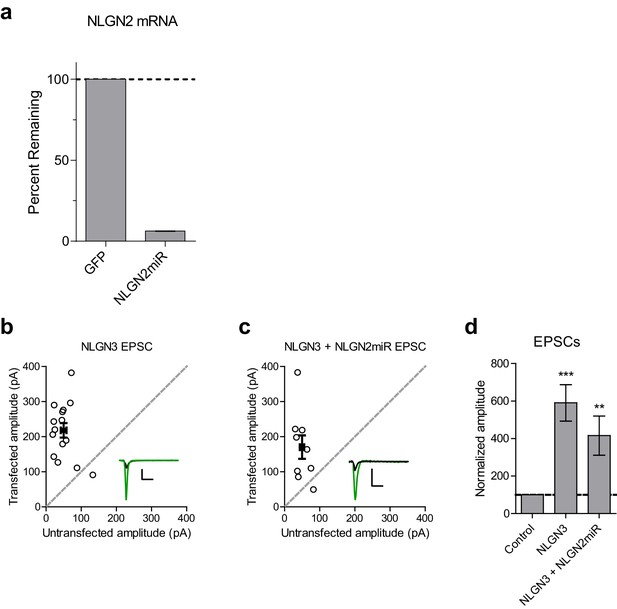
Validation of NLGN2 knockdown construct and EPSCs for NLGN3 overexpression.
(a) qRT-PCR bar graph shows mean ± s.e.m. of NLGN2 mRNA remaining following NLGN2miR transduction normalized to control GFP transduction (n = 2 technical replicates). (b) Scatter plot showing overexpression of NLGN3 enhances EPSCs compared to untransfected controls (***p=0.0002, n = 15). (c) Scatter plot showing overexpression of NLGN3 combined with NLGN2 knockdown enhances EPSCs (**p=0.0078, n = 9). (d) Summary graph of b and c. For panels b and c, open circles are individual pairs, filled circle is mean ± s.e.m. Black sample traces are control, green are transfected. Scale bars represent 100 pA and 50 ms. For panel d, summary graph plots mean transfected amplitude ± s.e.m, expressed as a percentage of control amplitude. Significance above each column represents pairwise comparison between transfected and untransfected cells
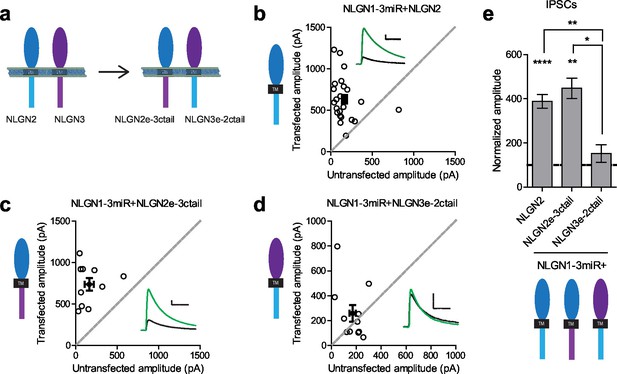
Difference between NLGN2 and 3 function at inhibitory synapses resides in the extracellular region.
(a) To determine where this functional difference between NLGN2 and 3 resides, we made chimeric constructs and expressed them on a reduced endogenous neuroligin background. Black TM is transmembrane domain. (b) Scatter plot showing expression of NLGN2 on a NLGN1-3miR knockdown background enhances IPSCs compared to untransfected controls (****p<0.0001, n = 25). (c) Scatter plot showing expression of a construct containing the NLGN2 extracellular region and NLGN3 cytoplasmic tail (NLGN2e-3ctail) on a NLGN1-3miR knockdown background enhanced IPSCs (**p=0.002, n = 10). (d) Scatter plot showing expression of a construct containing the NLGN3 extracellular region and NLGN2 cytoplasmic tail (NLGN3e-2ctail) on a NLGN1-3miR knockdown background failed to enhance IPSCs (p=0.5771, n = 11). (e) Summary graph of b-d. Effect of expressing NLGN3e-2ctail is significantly less than expression of NLGN2e-3ctail (*p=0.0124) or NLGN2 (**p=0.0067). For panels b-d, open circles are individual pairs, filled circle is mean ± s.e.m. Black sample traces are control, green are transfected. Scale bars represent 100 pA and 50 ms. For panel e, graph plots mean transfected amplitude ± s.e.m, expressed as a percentage of control amplitude. Significance above each column represents pairwise comparison between transfected and untransfected cells. See also Figure 2—figure supplement 1.
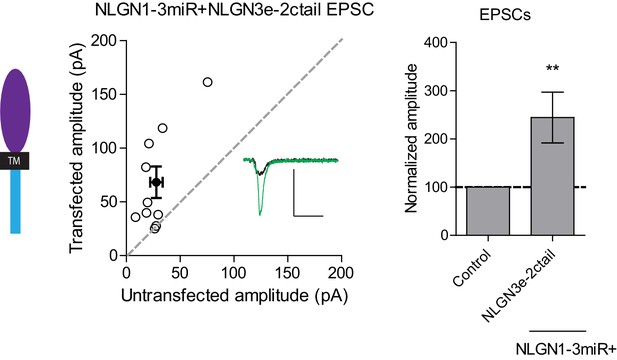
Distinct requirements for NLGN function between inhibitory and excitatory synapses.
While expression of a construct containing the NLGN3 extracellular region and NLGN2 cytoplasmic tail failed to rescue IPSCs, it was still able to enhance excitatory synaptic transmission (**p=0.0098, n = 10). Open circles are individual pairs, filled circle is mean ± s.e.m. Black sample trace is control, green is transfected. Scale bar represents 100 pA and 50 ms. Summary graph plots transfected amplitude normalized to control ± s.e.m. Significance represents pairwise comparison between transfected and untransfected cells.
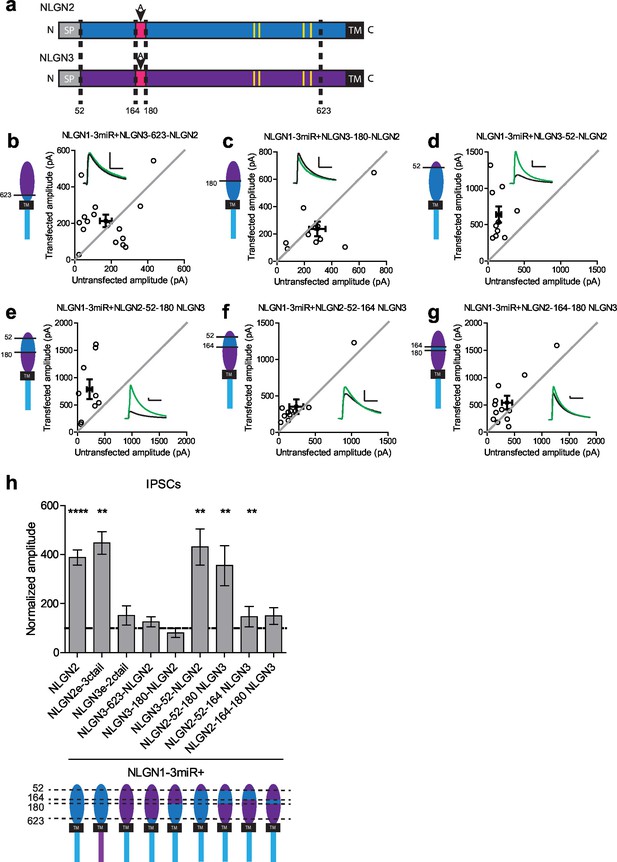
Identification of critical domain to confer NLGN function at inhibitory synapses.
(a) Schematic of NLGN2 and 3 showing positions of chimera. Yellow are critical dimerization residues, pink is splice site A, grey SP is signal peptide, and black TM is transmembrane domain. (b) Scatter plot showing that adding NLGN2 onto the NLGN3 extracellular region up to the 623rd residue fails to enhance IPSCs (p=0.4229, n = 16). (c) Scatter plot showing that adding up to the 180th residue also fails to enhance IPSCs (p=0.2754, n = 10). (d) Scatter plot showing that adding up to the 52nd residue is successful at conferring NLGN function at inhibitory synapses and enhancing IPSCs (**p=0.002, n = 10) (e) Expression of further chimeric constructs identifies a domain between residue 52–180 on NLGN2 that is sufficient to confer the ability to enhance IPSCs to NLGN3 (**p=0.0039, n = 9). Within this region, we also expressed only (f) the region between 52–164 (**p=0.0098, n = 10) or (g) residues corresponding to differences in splice site A (164–180) (p=0.0522, n = 12). (h) Summary graph. While the chimeric construct NLGN2-52-164-NLGN3 showed enhancement of IPSCs compared to untransfected control cells, the level of potentiation was significantly less than what we see with full-length NLGN2 (*p=0.0152). For panels b-g, open circles are individual pairs, filled circle is mean ± s.e.m. Black sample traces are control, green are transfected. Scale bars represent 100 pA and 50 ms. For panel h, graph plots mean transfected amplitude ± s.e.m, expressed as a percentage of control amplitude. Significance above each column represents pairwise comparison between transfected and untransfected cells. See also Figure 3—figure supplement 1.
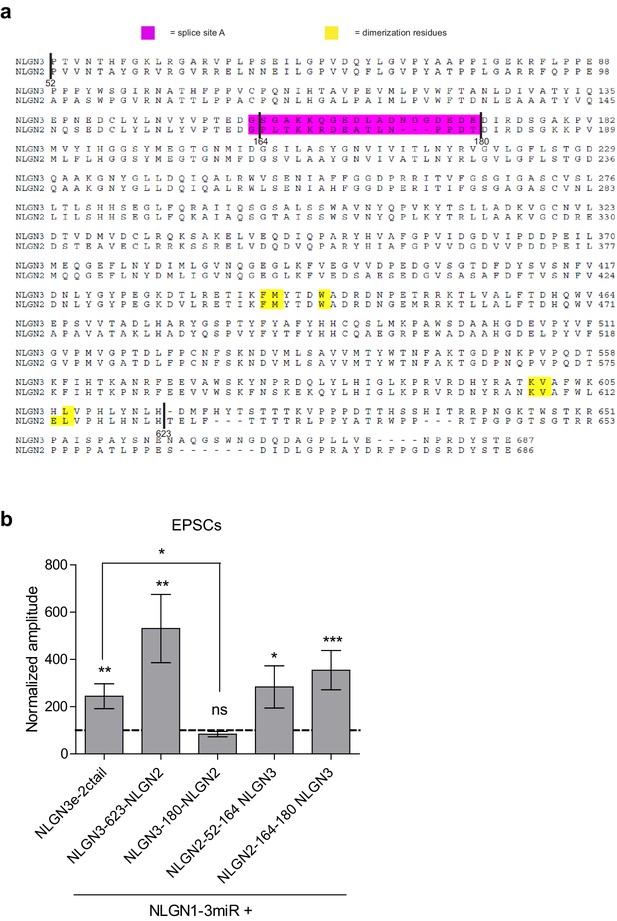
Chimeric extracellular mutants and effect on excitatory transmission.
(a) Alignment of the NLGN2 and NLGN3 extracellular regions. Sequence before residue 52 encompasses the highly dissimilar signal sequence and was not included in alignment. Highlighted in magenta is splice site A while key dimerization residues are highlighted in yellow. Relevant sites where chimeric constructs were made are identified in alignment. The first amino acid at the beginning of splice site A at residue 163 is conserved between NLGN2 and 3, therefore our chimera was made after this residue. (b) Excitatory responses of chimera that did not potentiate IPSCs. Constructs that did not potentiate IPSCs could still enhance EPSCs including: NLGN3e-2ctail (**p=0.0098, n = 10), NLGN3-623-NLGN2 (**p=0.0039, n = 9), NLGN2-52-164-NLGN3 (*p=0.0353, n = 14), NLGN2-164-180-NLGN3 (***p=0.0005, n = 13). The chimeric construct NLGN3-180-NLGN2 did not enhance EPSCs (p=0.3804, n = 12) and was significantly impaired compared to NLGN3e-2ctail (*p=0.0111). Summary graph plots transfected amplitude normalized to control ± s.e.m. Significance above each column represents pairwise comparison between transfected and untransfected cells.
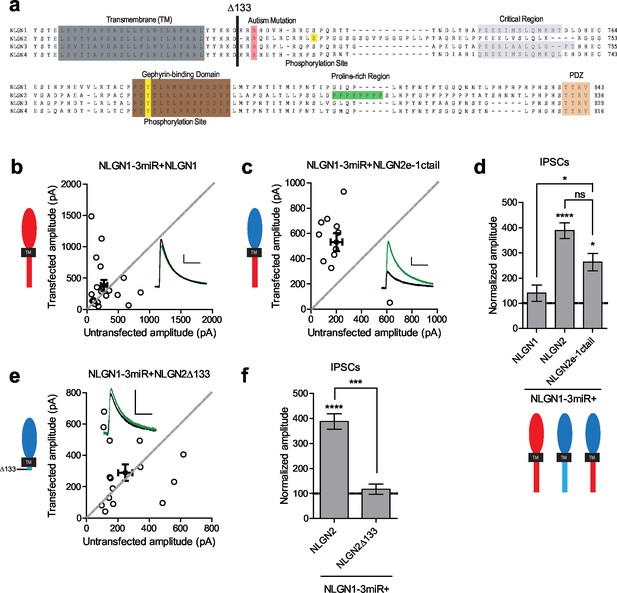
While NLGN c-tails do not confer specialization to inhibitory synapses, a c-tail is required for NLGN function.
(a) Alignment of NLGN1-4. Sequences correspond to mouse NLGN1, rat NLGN2, and human NLGN3 and NLGN4. Indicated at residue 133 is where most proximal c-tail truncation was performed. Highlighted in gray is the transmembrane domain. In light red is a conserved residue that is the site of an autism-associated mutation in NLGN4. Highlighted in light purple is the critical region, previously shown to be important for NLGN function at excitatory synapses but not inhibitory synapses (Shipman et al., 2011). Highlighted in brown is the gephyrin-binding domain. In green is the proline-rich region, and in light orange is the PDZ domain. Phosphorylation sites are highlighted in yellow. (b) Scatter plot showing expression of full-length NLGN1 on a NLGN1-3miR knockdown background failed to enhance inhibitory responses (p=0.6726, n = 19), (c) Scatter plot showing expression of a chimeric NLGN2 extracellular region with NLGN1 c-tail on a NLGN1-3miR knockdown background is able to enhance inhibitory responses (*p=0.0244, n = 11). (d) Summary graph. Effect of expressing NLGN2e-1ctail is significantly greater than with full-length NLGN1 (*p=0.0314). (e) Scatter plot showing expression of the c-tail truncation mutant NLGN2Δ133 on a reduced endogenous neuroligin background is unable to enhance inhibitory synaptic responses, indicating that a c-tail is required for NLGN function (p=0.6387, n = 15). (f) Summary graph of e. Effect of expressing NLGN2Δ133 is significantly less than with full-length NLGN2 (***p=0.0002). For panels b-c and e, open circles are individual pairs, filled circle is mean ± s.e.m. Black sample traces are control, green are transfected. Scale bars represent 100 pA and 50 ms. For panels d and f, graph plots transfected amplitude normalized to control ± s.e.m. Significance above each column represents pairwise comparison between transfected and untransfected cells. See also Figure 4—figure supplement 1.
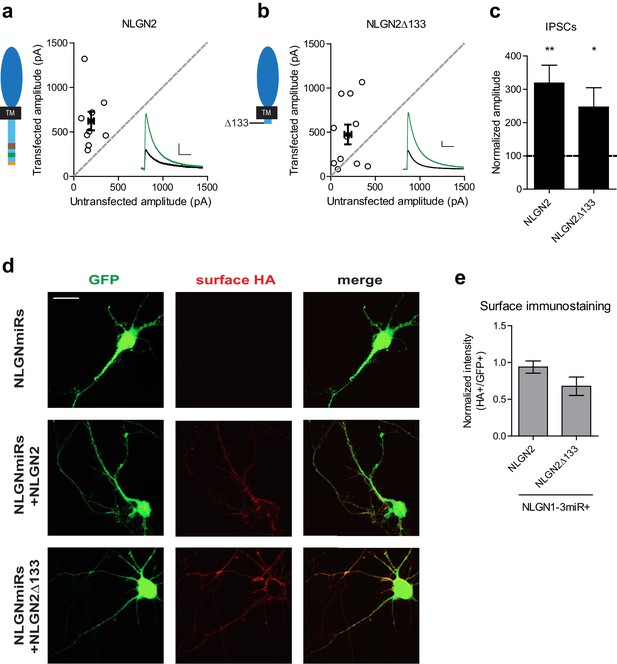
Further characterization of c-tail deletion mutant.
(a-b) Scatter plots showing overexpression on a wild-type background of (a) NLGN2 (**p=0.0039, n = 9) and (b) NLGN2Δ133 (*p=0.042, n = 11), both potentiate inhibitory responses. (c) Summary of a-b. (d) Immunostaining of dissociated hippocampal cultures transfected with NLGNmiRs-GFP alone or with either HA-tagged NLGN2 or HA-tagged NLGN2Δ133 and surface stained for HA. Scale bar represents 20 μm. (e) Summary of d. Graph plots intensity of HA signal normalized to GFP (p=0.189, n = 11 cells per group). Error bars are mean ± s.e.m. For panels a and b, open circles are individual pairs, filled circle is mean ± s.e.m. Black sample traces are control, green are transfected. Scale bars represent 100 pA and 50 ms. For panel c, graph plots transfected amplitude normalized to control ± s.e.m. Significance above each column represents pairwise comparison between transfected and untransfected cells.
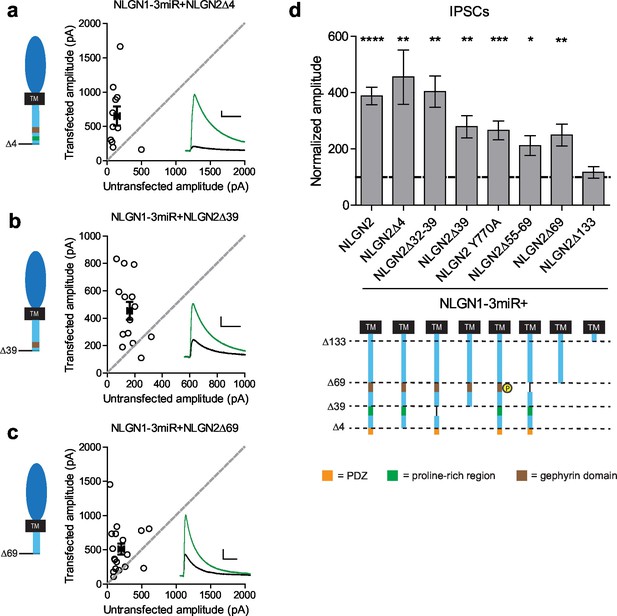
Previously identified domains not required for NLGN2 function.
(a–c) Scatter plots showing replacement of endogenous neuroligin with truncation mutants (a) lacking the PDZ domain (NLGN2Δ4) (**p=0.0098, n = 11), (b) truncation of all regions downstream of the proline rich region (NLGN2Δ39) (**p=0.0017, n = 14), or (c) truncation of all regions downstream of the gephyrin binding domain (NLGN2Δ69) (**p=0.0017, n = 18), is still able to potentiate inhibitory synaptic responses compared to paired neighboring control cells. (d) Summary graph. Shown are further mutants including deletion of only the proline rich region (NLGN2Δ32–39) (**p=0.002, n = 10), deletion of only the gephyrin binding domain (NLGN2Δ55–69) (*p=0.0273, n = 10) or expression of a phospho-mimic point mutant that has been shown to disrupt gephyrin binding (NLGN2 Y770A) (***p=0.0005, n = 17), which all still potentiate inhibitory responses. For panels a-c, open circles are individual pairs, filled circle is mean ± s.e.m. Black sample traces are control, green are transfected. Scale bars represent 100 pA and 50 ms. For panel d, graph plots transfected amplitude normalized to control ± s.e.m. Significance above each column represents pairwise comparison between transfected and untransfected cells. See also Figure 5—figure supplement 1.
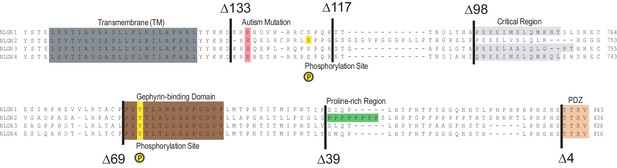
More detailed NLGN alignment showing where all truncations and point mutations were made.
Highlighted in gray is the transmembrane domain. In light red is a conserved residue that is a site of an autism-associated mutation in NLGN4. Highlighted in light purple is the critical region, previously shown to be important for NLGN function at excitatory synapses but not inhibitory synapses (Shipman et al., 2011). Highlighted in brown is the gephyrin-binding domain. In green is the proline-rich region, and in light orange is the PDZ domain. Phosphorylation sites are highlighted in yellow.
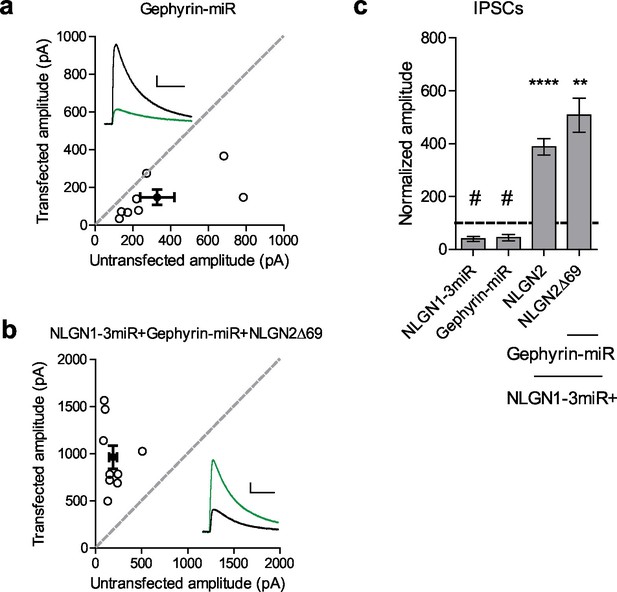
Gephyrin-independent NLGN2 function.
(a) Scatter plot showing overexpression of a Gephyrin-miR construct to knockdown gephyrin expression significantly reduces inhibitory synaptic responses (#p=0.0156, n = 8). (b) Scatter plot showing replacement of endogenous neuroligin with a truncation mutant lacking all known domains of neuroligin 2 (NLGN2Δ69) on a gephyrin knockdown background is still able to potentiate inhibitory synaptic responses, indicating that neuroligin 2 is able to function independently of gephyrin (**p=0.0039, n = 9). (c) Summary graph. Asterisks indicate significant enhancement compared to control cells where **p<0.01 and ***p<0.001, while hash sign indicates significant depression compared to control cells where #p<0.05. For panels a and b, open circles are individual pairs, filled circle is mean ± s.e.m. Black sample traces are control, green are transfected. Scale bars represent 100 pA and 50 ms. For panel c, graph plots transfected amplitude normalized to control ± s.e.m. Significance above each column represents pairwise comparison between transfected and untransfected cells. See also Figure 6—figure supplement 1.
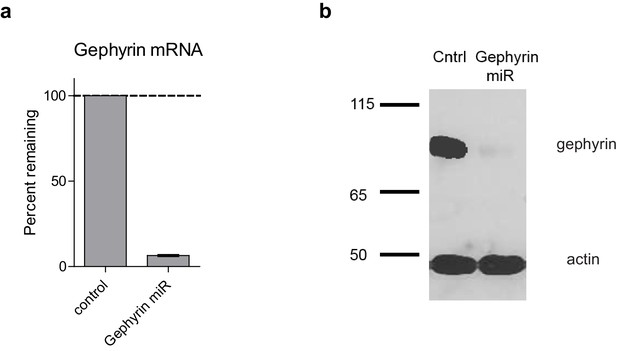
Characterization of Gephyrin knockdown construct.
(a) Graph shows mean ± s.e.m. of gephyrin mRNA remaining following Gephyrin-miR transduction normalized to control (n = 3 technical replicates). (b) Western blot of lysates taken from dissociated hippocampal cultures following Gephyrin-miR transduction compared to control uninfected neurons. Shown is representative image of 6 technical replicates. Top band is gephyrin, lower band is actin. Size in kDa.
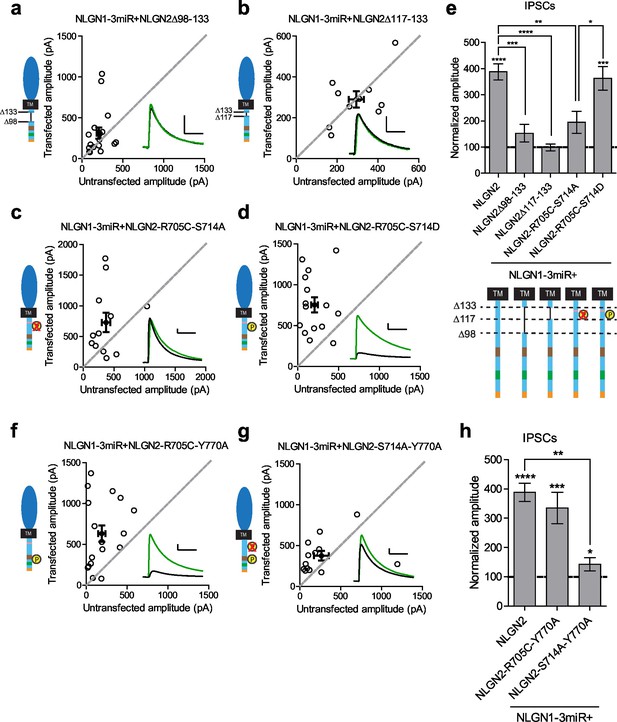
Identification of critical residues in NLGN2 c-tail.
(a) Scatter plot showing deletion of a region from the most proximal c-tail truncation (NLGN2Δ98–133) failed to enhance inhibitory responses (p=0.1698, n = 17). (b) Further refining this area of interest, NLGN2Δ117–133 still failed to enhance inhibitory responses (p=0.6953, n = 10). (c) Scatter plot showing double point mutant with autism-associated mutation at R705 and phospho-null mutation at S714 (NLGN2-R705C-S714A) also showed deficits in inhibitory transmission (p=0.0522, n = 12). (d) Scatter plot showing double point mutant with autism-associated mutation at R705 and phospho-mimic mutation at S714 (NLGN2-R705C-S714D) displayed normal enhancement of IPSCs (***p=0.0002, n = 15). (e) Summary graph. Expression of full-length NLGN2 results in greater enhancement of IPSCs compared to NLGN2Δ98–133 (***p=0.0001), NLGN2Δ117–133 (****p<0.0001), and NLGN2-R705C-S714A (**p=0.0061). Expression of NLGN2-R705C-S714D results in greater enhancement of IPSCs compared to NLGN2-R705C-S714A (*p = 0.0429). (f) Scatter plot showing double point mutant with autism-associated mutation at R705 and a phospho-mimic point mutant shown to disrupt gephyrin binding at Y770 is still able to enhance inhibitory responses (***p=0.0005, n = 17). (g) Scatter plot showing double point mutant with phospho-null mutation at S714 and a phospho-mimic point mutant shown to disrupt gephyrin binding at Y770 exhibited modest enhancement of IPSCs (*p=0.0274, n = 13). (h) Summary graph. While the double mutant NLGN2-S714A-Y770A showed enhancement of IPSCs compared to untransfected control cells, the level of potentiation was far less than what we see with full-length NLGN2 (**p=0.0089). For panels a-d, and f-g open circles are individual pairs, filled circle is mean ± s.e.m. Black sample traces are control, green are transfected. Scale bars represent 100 pA and 50 ms. For panel e and h, bar graph plots transfected amplitude normalized to control ± s.e.m. Significance above each column represents pairwise comparison between transfected and untransfected cells. See also Figure 7—figure supplement 1.
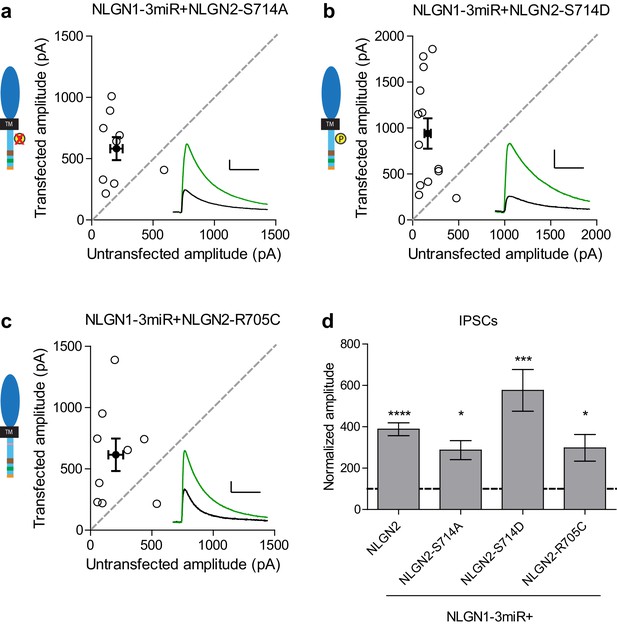
Individual c-tail point mutations do not affect NLGN2 function.
(a–c) Scatter plots showing replacement of endogenous neuroligin with individual mutants of either (a) phospho-null mutation (*p=0.0195, n = 9), (b) phospho-mimic mutation (***p=0.0007, n = 13), or (c) autism-associated mutation (*p=0.0391, n = 9) still potentiates IPSCs. (d) Summary graph. For panels a-c, open circles are individual pairs, filled circle is mean ± s.e.m. Black sample traces are control, green are transfected. Scale bars represent 100 pA and 50 ms. For panel d, bar graph plots transfected amplitude normalized to control ± s.e.m. Significance above each column represents pairwise comparison between transfected and untransfected cells.
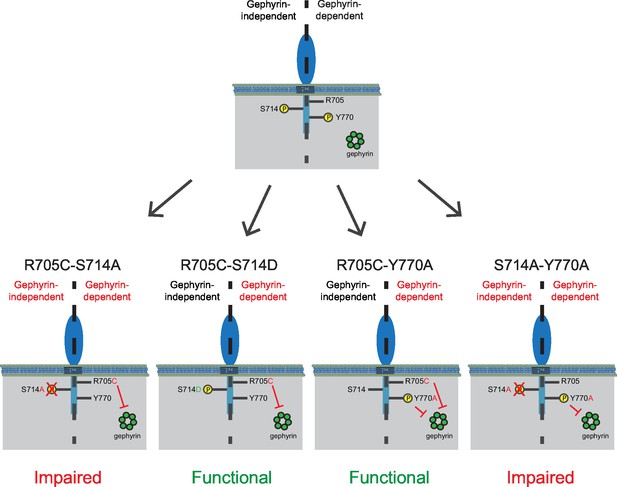
Separate gephyrin-dependent and gephyrin-independent mechanisms for neuroligin function at inhibitory synapses.
We propose separate gephyrin-dependent and gephyrin-independent mechanisms for NLGN function at inhibitory synapses. In our NLGN2-R705C-S714A manipulation, we block the gephyrin-independent pathway by preventing phosphorylation at the S714 residue, and also block the gephyrin-dependent pathway by the autism-associated mutation at R705. Therefore, the resulting effect we see is impaired function at inhibitory synapses. In our NLGN2-R705C-S714D manipulation, we facilitate the gephyrin-independent pathway by mimicking phosphorylation at the S714 residue, while the gephyrin-dependent pathway is blocked by the autism-associated mutation at R705. The resulting effect we see is intact function at inhibitory synapses. In our NLGN2-R705C-Y770A manipulation, we only block the gephyrin-dependent pathway with the autism-associated mutation at R705 and a mutation at Y770 that blocks gephyrin-binding. The resulting effect we see is intact function at inhibitory synapses. In our NLGN2-S714A-Y770A manipulation, we block the gephyrin-independent pathway by preventing phosphorylation at the S714 residue, and also block the gephyrin-dependent pathway with the mutation at Y770 that blocks gephyrin-binding. Therefore, the resulting effect we see is impaired function at inhibitory synapses. Green is gephyrin. Yellow residues are phosphorylation sites. The Y770A point mutant has been previously characterized and shown to function as a phospho-mimic mutation that blocks gephyrin binding (Giannone et al., 2013).
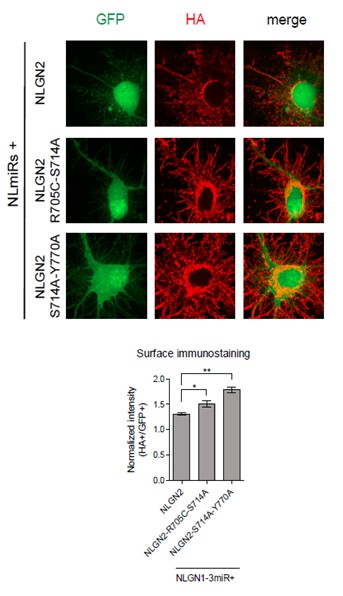
Dissociated hippocampal rat cultures were prepared at E18.5 and transfected with NLmiRs-GFP and either NLGN2, NLGN2-R705C-S714A, or NLGN2-S714A-Y770A at DIV 8.
After 1 week cells were stained for surface HA (Abcam ab9110, 1:200). HA signal was normalized to GFP. (NLGN2 n=5 cells, NLGN2-R705C-S714A n=7 cells, NLGN2-S714A-Y770A n=7 cells; *p=0.0177, **p=0.0025).
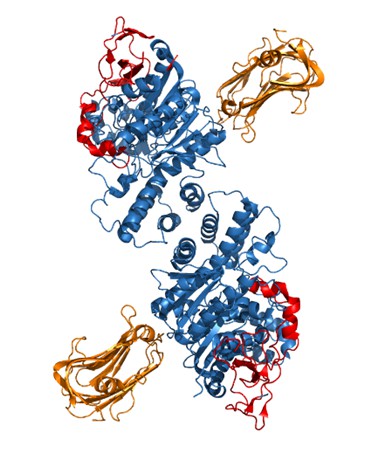
Crystal structure of NLGN2 dimer (in blue) aligned with Neurexin1β structure (in orange).
In red is the critical extracellular domain in NLGN2 identified in our study.
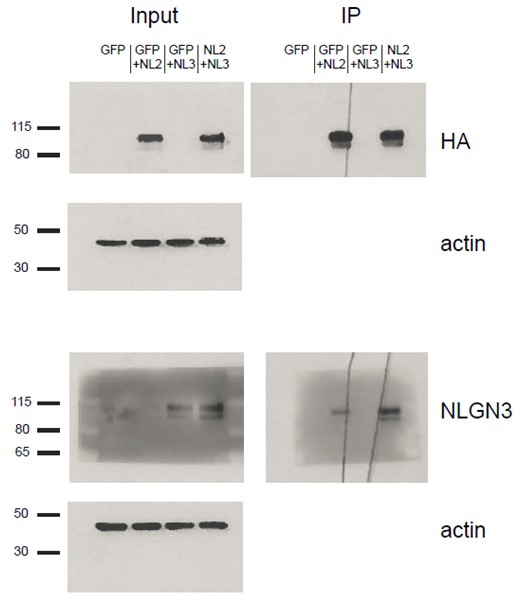
HEK293T cells were transfected with either GFP, GFP+NLGN2, GFP+NLGN3, or NLGN2+NLGN3.
After 2 days lysates were harvested and incubated in HA-conjugated agarose beads (Sigma-Aldrich, Saint Louis, MO) to immunoprecipitate HA-tagged protein complexes. Samples were run on a 4–12% Bis-Tris gel and probed for HA (Santa Cruz 1:2000), NLGN3 (Synaptic Systems 1:1000), and actin (Millipore C4 1:5000). Input samples are 3% of IP samples. Blots are representative of at least 2 experimental and technical replicates. Size indicated on left in kDa.
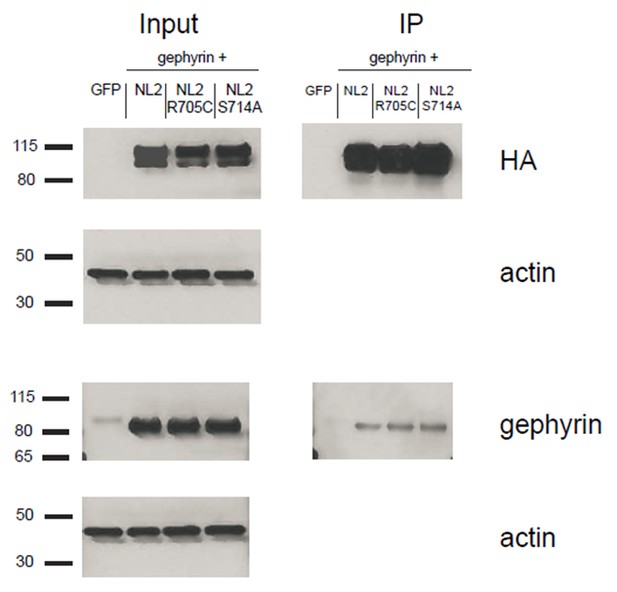
HEK293T cells were transfected with either GFP, NLGN2+gephyrin, NLGN2R705C+gephryin, or NLGN2S714A+gephyrin.
After 2 days lysates were harvested and incubated in HA-conjugated agarose beads (Sigma) to immunoprecipitate HA-tagged protein complexes. Samples were run on a 4–12% Bis-Tris gel and probed for HA (Santa Cruz 1:2000), gephyrin (Synaptic Systems 1:5000), and actin (Millipore C4 1:5000). Input samples are 3% of IP samples. Blots are representative of at least 2 technical replicates. Size indicated on left in kDa.
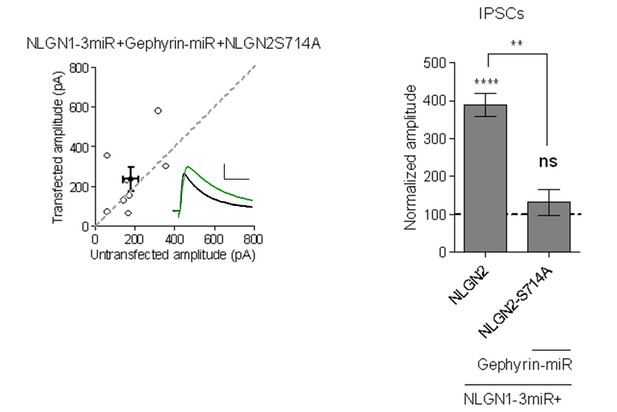
Scatterplot and bar graph showing expression of NLGN2S714A along with NLGN1-3miR and Gephyrin-miR fails to enhance inhibitory responses and is significantly different than expression of full-length NLGN2 on the NLGN1-3miR background (n = 8, **p=0.0023, ****p<0.0001).
Scale bar represents 25pA and 50ms.





