Pathogen effectors and plant immunity determine specialization of the blast fungus to rice subspecies
Figures
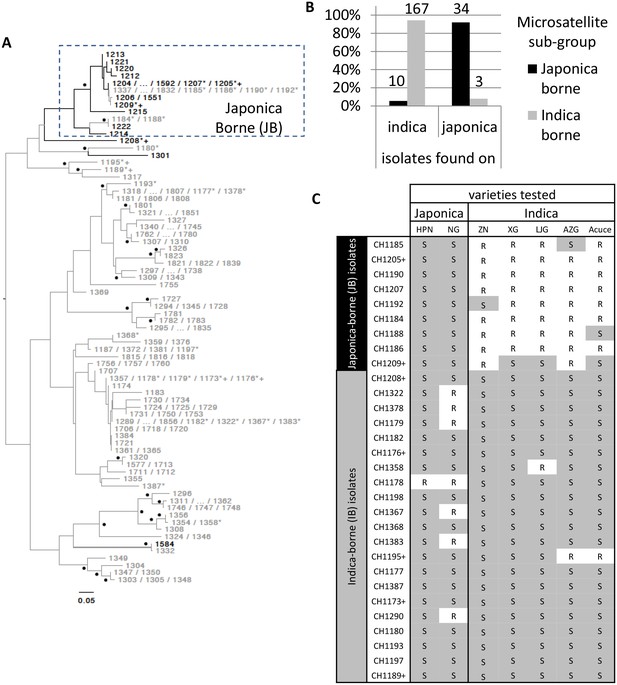
Variability in microsatellite genotype and pathogenicity phenotype of M. oryzae isolates harvested on indica and japonica rice grown in Yuanyang terraces.
(A) Midpoint rooted neighbor-joining dendrogram representing the proportion of shared microsatellite alleles among multilocus genotypes. Two hundred fourteen isolates (the prefix ‘CH’ visible in C was removed from A for clarity) were genotyped using 13 microsatellites. Only one representative of multilocus genotypes repeated multiple times was kept, and for each repeated multilocus genotype the corresponding isolates are listed at the tip of a branch (74 unique multilocus genotypes in total). Bootstrap supports are indicated by a black dot when >40% (1000 resamplings). Isolates harvested on japonica and indica rice are indicated in black and grey respectively. Six isolates (i.e. CH1180, CH1189, CH1195, and CH1317, collected on indica; CH1208 and CH1301, collected on japonica) that show in the dendrogram an intermediate position, were assigned to the IB group because of their pathogenicity phenotypes and their separation from the other JB genotypes in the DAPC (Figure 1—figure supplement 1). The isolates selected for (C) and Figure 3 are marked by a ‘*’ and a ‘+’ respectively. The cluster of Japonica-borne isolates (abbreviated JB; within the square with dashed lines) was defined following Discriminant Analysis of Principal Components; the remaining samples represent the cluster of Indica-borne (IB) isolates. (B) Distribution of the two clusters identified based on microsatellite variation (japonica borne ‘JB’ and indica borne ‘IB’) on japonica and indica hosts in the Yuanyang terraces. The fact that the distributions are largely non-overlapping (the JB and IB clusters are mostly found on Japonica and Indica hosts, respectively) suggests local adaptation of the pathogens to their respective hosts. The numbers of isolates are indicated above the bars. (C) Pathogenicity profiles of 30 isolates on two japonica and five indica varieties (‘R’ and ‘S’ stand for resistance and susceptibility, respectively). The 30 representative isolates were selected from the analysis presented in (A) and inoculated onto seven major rice varieties grown in Yuanyang (HPN: Huang Pi Nuo; NG: Nuo Guo; ZN: Zi Nuo; XG: Xiao Gu; LJG: Li Jiao Gu; AZG: Ai Zhe Gu; Acuce). HPN, NG and ZN are all glutinous rice varieties. The isolates marked with a ‘+’ in (A) and (C) are those used in Figure 3; all isolates included in (C) are marked with a ‘*’ in (A). The qualitative analysis of symptoms presented here suggests that japonica-borne (JB) isolates cannot attack indica rice whereas indica-borne (IB) isolates can attack japonica.
-
Figure 1—source data 1
The data relates to Figure 1.
For each of the 215 Magnaporthe oryzae isolate, the name of the variety on which it was collected is indicated and the rice subspecies is indicated (indica or japonica). The ‘pyrm’ columns represent microsatellites names from Saleh et al. (2014). Other informations like GPS position, altitude and town where the isolates were collected are also indicated.
- https://doi.org/10.7554/eLife.19377.004
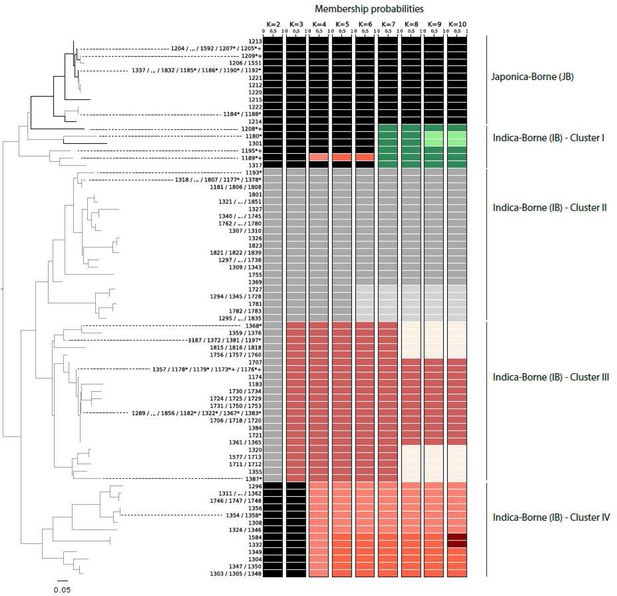
Neighbor-joining tree representing the genetic distance (in terms of proportion of shared alleles) between the 74 unique microsatellite genotypes characterized in Yuanyang (left) and patterns of memberships in K = 2 to K = 10 clusters as inferred using DAPC (right).
https://doi.org/10.7554/eLife.19377.005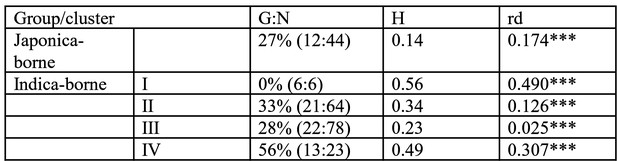
Summary statistics of genetic variability in the four clusters of Magnaporthe oryzae identified using Discriminant Analysis of Principal Components and neighbor-joining analysis of genetic distance (see Figure 1—figure supplement 1).
G:N is the number of unique genotypes, divided by the total number of genotypes. rd is the standardized index of association a measure of multilocus linkage disequilibrium, computed on clone corrected datasets (i.e. keeping a single representative for each multilocus genotypes represented multiple times) ; significance was determined using 1000 randomizations to simulate random mating. H is the unbiased gene diversity, averaged across loci. ***p<0.001.
Midpoint rooted neighbor-joining dendrogram representing the proportion of shared microsatellite alleles among the 214 multilocus genotypes originating from Yuanyang terraces.
The topology is different from Figure 1A, which was based on unique multilocus genotype only, while this analysis was based on the full set of isolates. Isolates were colored according to their collection year (left-hand side panel) or the collection altitude (right-hand side panel), showing no clear pattern of clustering of isolates according to these factors.
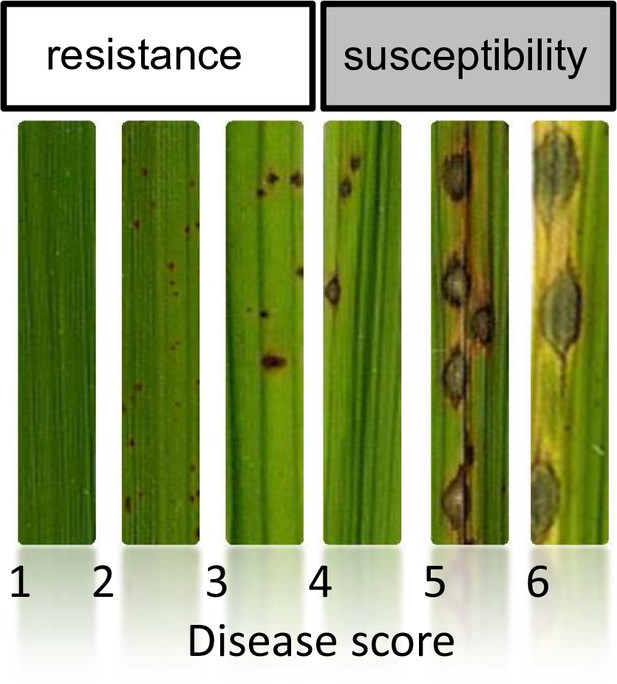
Scale used for scoring global incompatibility/compatibility (Resistance/Susceptibility).
The small brown lesions are indicative of infection sites were the fungus was stopped (Hypersensitive-response) whereas large and greyish lesions represent successful penetration and multiplication of the pathogen. This notation system is the same than in Gallet et al (Gallet et al., 2016).
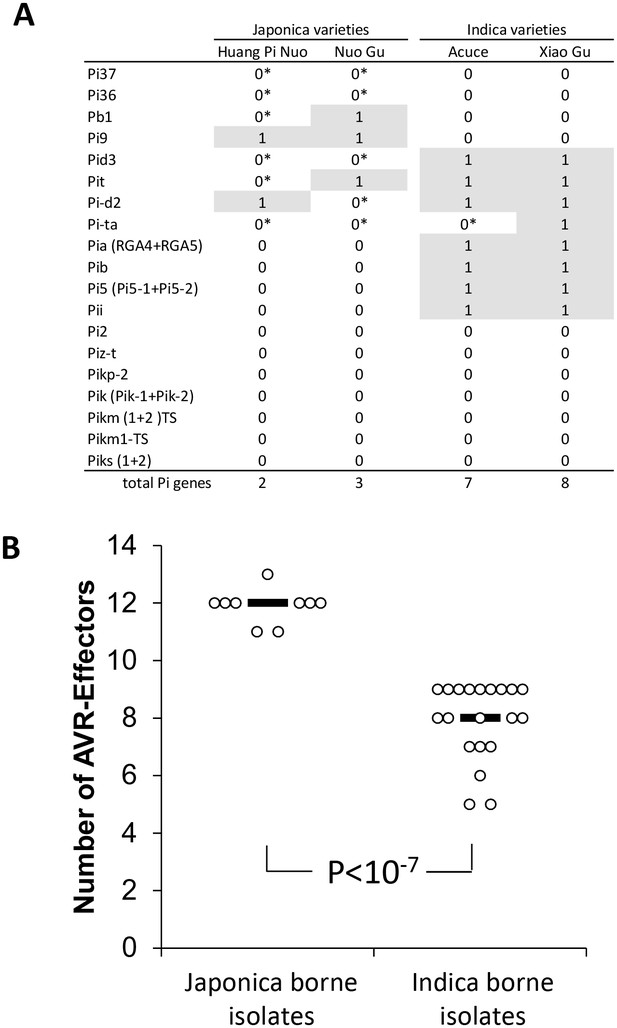
Evaluation of rice Pi and blast Avr-effectors genes in Yuanyang rice varieties and M. oryzae isolates.
(A) Presence (1)/absence (0) of 19 cloned Pi gene based on sequence analysis. Pair-end reads from the four varieties were produced (53 to 58 million reads) by whole-genome sequencing. The reads (~150 nucleotides) were mapped on the corresponding Pi sequences using SOAPaligner (http://soap.genomics.org.cn/soapaligner.html) and two mismatches were allowed. Some genes (noted 0*) were present but contained a premature stop codon or were not functional according to inoculation tests (Figure 2—figure supplement 2). The sequences used are listed in Figure 2—figure supplement 1. Note that when two genes are required for resistance (Pia, Pi5, Pik, Pik-s and Pik-m), the sequences of both genes were analyzed. (B) The number of Avr function of 14 effectors was measured (Figure 2—figure supplement 4) in the JB and IB groups of 30 isolates defined in Figure 1C. Each dot represents an isolate and the median (black bar) is indicated. The average values between JB and IB isolates are significantly different (p<10−7; t-test).
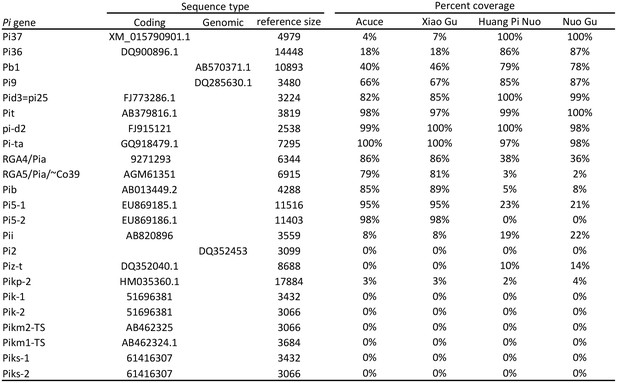
Estimation of the presence of Pi genes by mapping reads from whole-genome sequencing on the corresponding Pi gene sequence.
The pair-end reads from four species were aligned to the reference library using SOAPaligner (http://soap.genomics.org.cn/soapaligner.html), respectively, and just two mismatches were allowed. The parameters were “soap -a HH-8_AC_left_reads.fq -b HH-8_AC_right_reads.fq -D ref_library.fasta -o HH-8_AC.PE_align −2 HH-8_AC.SE_align -v 2 m 250 -x 450 r 2 p 20” (for HH-8_AC, other were same). The reads which uniquely mapped to a gene were used to calculate coverage. The list below provides the reads’ statistics of each genome.
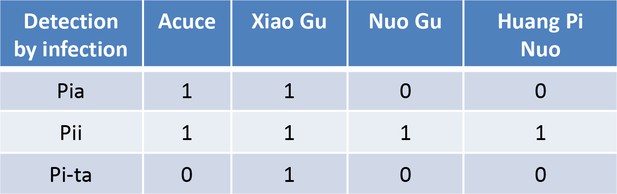
Estimation of the presence of the indicated Pi genes by inoculation with GUY11 transformed with the cognate Avr-Effector. “1” is presence (plant is resistant), “0” is absence of detection (plant is susceptible).
https://doi.org/10.7554/eLife.19377.011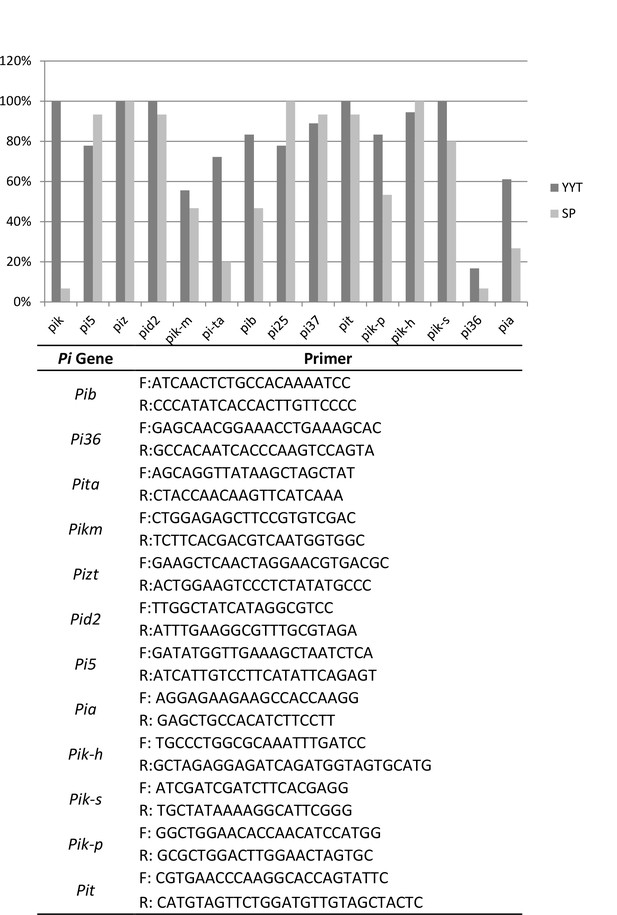
Estimation of the frequency of Pi genes in 18 (other than those listed in Figure 2A) indica, traditional varieties from Yuanyang terraces (YYT) and in 15 modern, improved indica varieties from the Shipping county (SP; Yunnan, China) used as a local geographical control.
The presence of the Pi gene was estimated using PCR primers listed.
Estimation of Avr-Effector complement using rice differential lines in 30 selected isolates from Yuanyang (these isolates are indicated by a ‘*’ on Figure 1A showing neutral diversity).
The Resistant (R) and susceptible (S) classes were established using the scale described in Figure 1—figure supplement 4. The isolates indicated with a ‘+’ were used for pathogenicity tests in Figure 4.
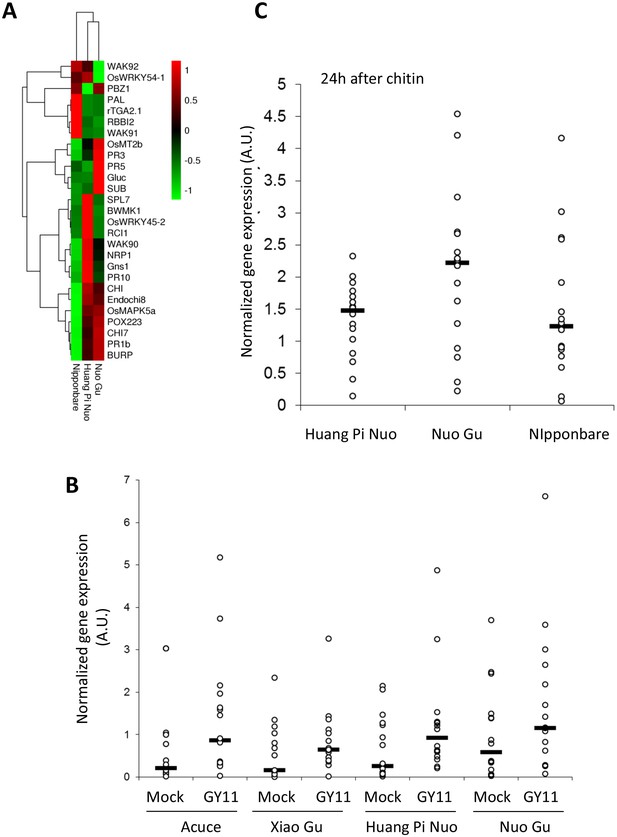
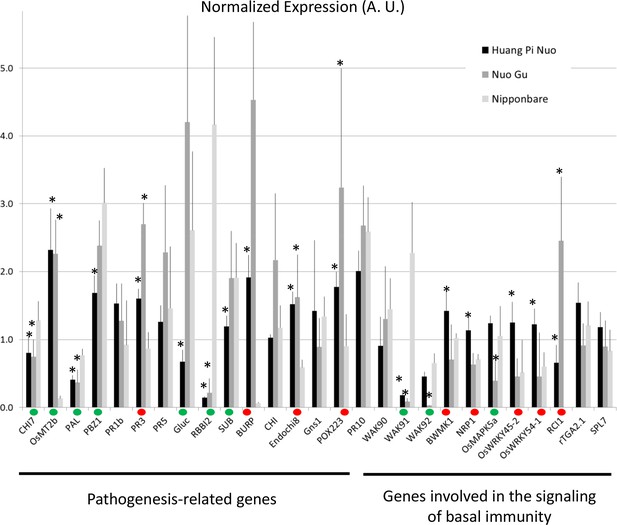
Constitutive and inducible defense in Yuanyang rice varieties.
The expression of defense-related genes (see Figure 3—figure supplement 3) in indica (Acuce, Xiao Gu) and japonica (Huang Pi Nuo and Nuo Gu) varieties grown in Yuanyang as well as the japonica Nipponbare variety was measured by RT-qPCR. In order to make genes with different expression levels comparable, the different values obtained for each gene were normalized by the average value of the considered gene across all measures. (A) The constitutive expression of 16 Pathogenesis-related genes and 11 genes involved in basal immunity signaling was measured by RT-qPCR. The mean values from four biological replicates was normalized and used for hierarchical clustering using hcluster algorithm (www.omicshare.com/tools). The corresponding mean, SD and statistical tests can be found in Figure 3—figure supplement 4. (B) The two major indica (Acuce and Xiao Gu) and japonica varieties found in Yuanyang (Huang Pi Nuo and Nuo Gu) were inoculated with the virulent isolate Guy11 or mock treated. The expression of 16 Pathogenesis Related-genes was measured and the mean from four biological replicates was calculated. Each dot represents the average expression value from 16 defense genes. The black bar represents the median value. The corresponding mean, SD and statistical tests can be found in Figure 3—figure supplement 5. (C) The expression of 16 Pathogenesis-related genes was measured 24 hr after chitin (100 μg/mL) treatment. The mean value from four biological replicates was calculated and each dot represents this value for one gene. The corresponding mean, SD and statistical tests can be found in Figure 3—figure supplement 7.
-
Figure 3—source data 1
The data relates to Figure 3.
The expression values of defense-related genes (columns) normalized by the Actin gene are given. All values were also normalized using the mean of each gene in order to make all genes comparable with each other. In the first columns, the first element represents the treatment (chi=chitin), the second the variety, the third the time after treatment and the last the replicate number.
- https://doi.org/10.7554/eLife.19377.015
-
Figure 3—source data 2
The data relates to Figure 3—figure supplement 2.
Columns 2-5 represent susceptibility to the corresponding M. oryzae isolate (CD203, CM28, CL26, and GY11). The data are then normalized by the value in the Maratelli (used as universal susceptible control) variety to allow comparisons between isolates.
- https://doi.org/10.7554/eLife.19377.016
-
Figure 3—source data 3
The data relates to Figure 3—figure supplement 4.
The expression of defense-related genes was evaluated by RT-qPCR and the data normalized by the actin gene is given. Each condition was replicated 3–4 times (column C). AC=Acuce, XG=Xiao Gu, HPN=Huang Pi Nuo, NG=Nuo Gu are rice varities. CL26 (A) and CM28 (B) are isolates of Magnaporthe oryzae isolates.
- https://doi.org/10.7554/eLife.19377.017
Examples of symptoms on Huang Pi Nuo.
The red and blue arrows indicate typical susceptible and resistance symptoms respectively. The yellow arrow shows a susceptible lesion with surrounding browning, a phenomenon associated with a local resistance response (Hayashi et al., 2016).
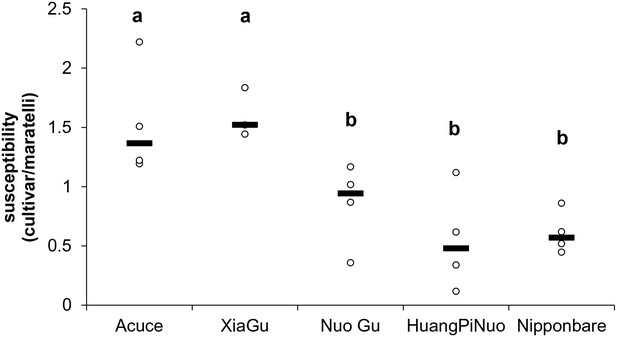
Average susceptibility of Yuanyang terraces varieties.
Four multi-virulent isolates (CD203, GUY11, CL26 and CM28 [Vergne et al., 2010]) were used to evaluate susceptibility in the absence of major Avr/Pi interactions. The number of susceptible lesions/unit surface was measured for each isolate X variety combination in four replicates of 6 leaves. The median value of four measures is indicated. Basal immunity can be extrapolated considering that it is inversely correlated to global susceptibility. Acuce, Xiao Gu are indica varieties. Huang Pi Nuo, Nuo Gu and Nipponbare are japonica varieties. Letters above indicate significantly different groups of values based on T-test (p<0.05).

Accessions and names of genes used for expression analysis.
The primers can be found in (Vergne et al., 2010) and (Delteil et al., 2012).
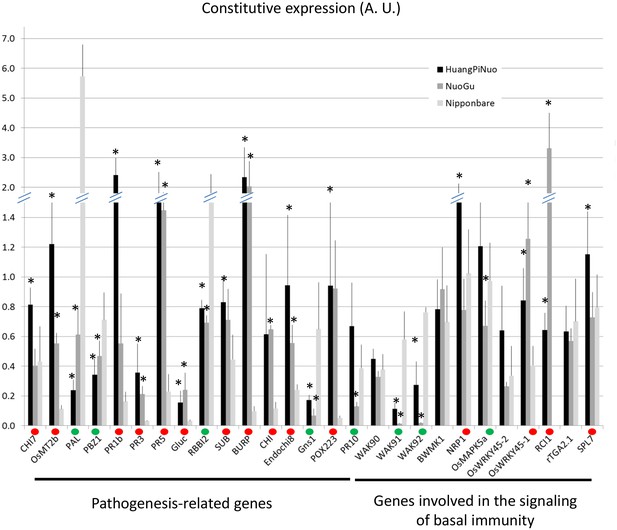
Constitutive expression of defense-related genes in Yuanyang japonica and Nipponbare varieties.
The japonica variety Nipponbare with high basal immunity was compared to the Yuanyang japonica varieties Huang Pi Nuo and Nuo Gu. The constitutive expression of 16 pathogenesis-related genes and 11 genes involved in basal immunity signaling was measured by RT-qPCR. In order to compare all genes together, all values for one gene were normalized with the average value of the considered gene for all measures. The values are the means and SD from four biological replicates. The ‘*’ denote statistical differences (t-test; p<0.05) between Nipponbare and the considered variety. A red and green dot indicates that Nipponbare expression is lower and higher than either Huang Pi Nuo or Nuo Gu respectively.
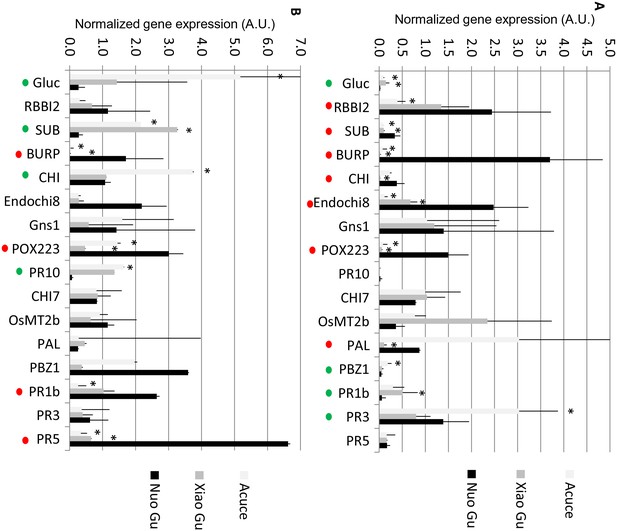
Constitutive and fungal-induced expression of pathogenesis-related genes in Yuanyang japonica and indica varieties.
The expression of 16 pathogenesis-related genes was measured by RT-qPCR in the absence of treatment. (A) or 24 after inoculation by M. oryzae (isolate Guy11). In order to compare all genes together, all values for one gene were normalized with the average value of the considered gene for all measures. The values are the means and SD from four biological replicates. The japonica variety Nuo Gu was compared to the Yuanyang indica varieties Acuce and Xiao Gu. The ‘*’ denotes statistical differences (t-test; p<0.05) between Nuo Gu and the considered variety. A red and green dot indicates that Nuo Gu expression is higher and lower than either Acuce or Xiao Gu.
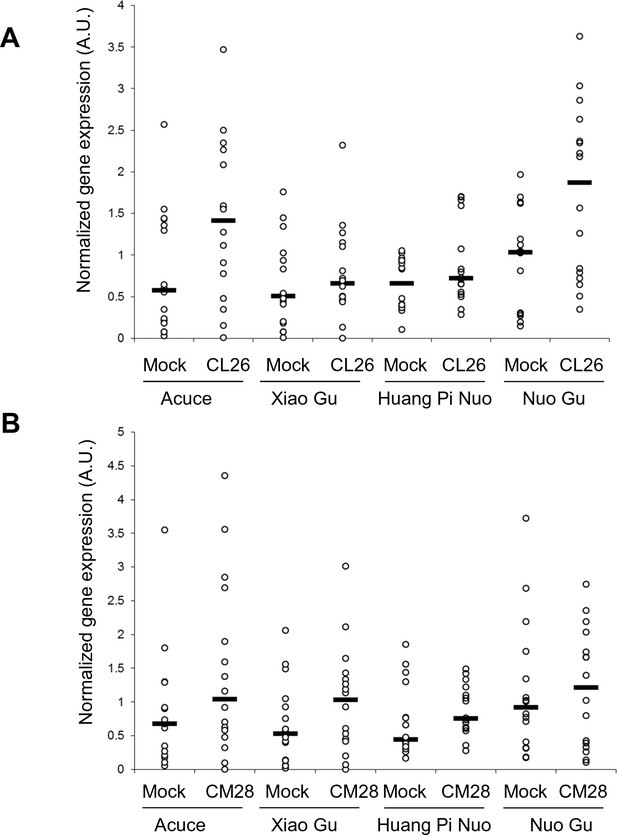
Constitutive and fungal-induced expression of pathogenesis-related genes in Yuanyang japonica and indica varieties.
The two major indica (Acuce and Xiao Gu) and japonica (Huang Pi Nuo and Nuo Gu) varieties found in Yuanyang were mock treated or inoculated with the virulent isolates CL26 (A) and CM28 (B). The expression of Pathogenesis-Related genes was measured by RT-qPCR and the mean from four biological replicates was calculated. Each dot represents the average expression value of 16 Pathogenesis Related-genes. The black bar represents the median value.
Chitin-induced expression of defense-related genes in Yuanyang japonica and Nipponbare varieties.
The japonica variety Nipponbare with high basal immunity was compared to the Yuanyang japonica varieties Huang Pi Nuo and Nuo Gu. The expression of 16 pathogenesis-related genes and 11 genes involved in basal immunity signaling was measured by RT-qPCR 24 hr after chitin treatment (100 ug/mL). In order to compare all genes together, all values for one gene were normalized with the average value of the considered gene for all measures. The values are the means and SD from four biological replicates. The ‘*’ denote statistical differences (t-test; p<0.05) between Nipponbare and the considered variety. A red and green dot indicates that Nipponbare expression is lower and higher than either Huang Pi Nuo or Nuo Gu respectively.
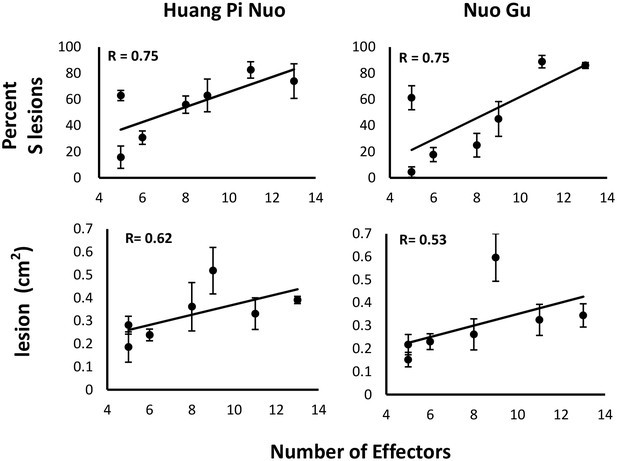
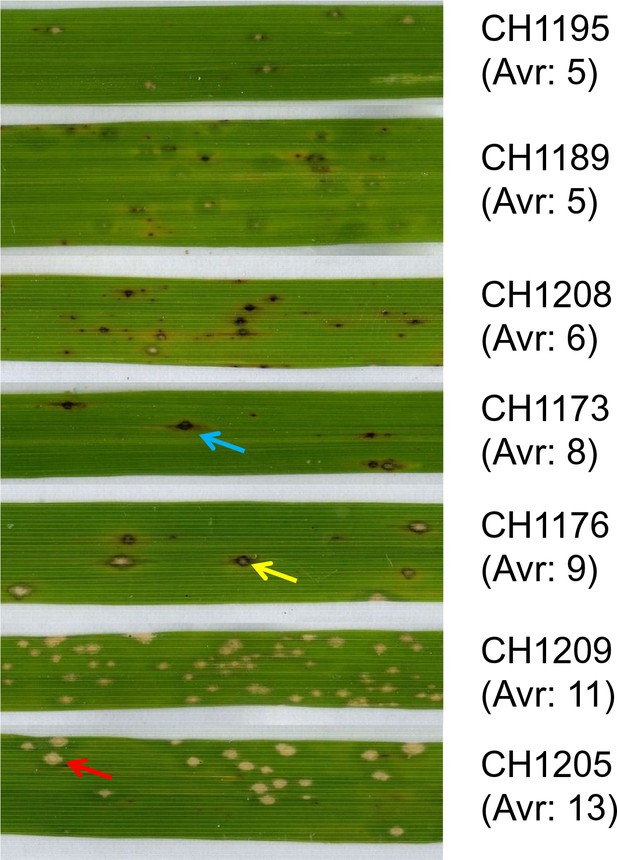
Effector complement and virulence of Yuanyang isolates.
Seven isolates with Avr-Effector number ranging from 5 to 13 (see isolates marked with ‘+’ in Figure 1) were selected and inoculated onto japonica varieties Huang Pi Nuo and Nuo Gu. The two quantitative components of virulence (lesion surface and percentage of susceptible lesions over total lesion number) are indicated as means and standard deviation from three biological replicates (each replicate included six independent plants).
-
Figure 4—source data 1
The data relates to Figure 4.
The file presents the data of inoculation of the Huang Pi Nuo (HPN) and Nuo Gu (NG) by different isolates of M. oryzae (strains) for each of which the number of AVR-effector is indicated. The first two spreadsheets are surfaces of susceptible lesions with HPN and NG. The last two spreadsheets are the data for calculating the percentage of susceptible lesions. Data of resistant (R) and susceptible (S) lesions are provided and used for calculating the total number of lesions.
- https://doi.org/10.7554/eLife.19377.026
-
Figure 4—source data 2
The data relates to Figure 4—figure supplement 1.
The values represent the number of lesions per unit surface for each of the replicates. The first lane represents the isolate used for inoculating the Nipponbare (Nip) or CEBiP (cebip) mutant.
- https://doi.org/10.7554/eLife.19377.027
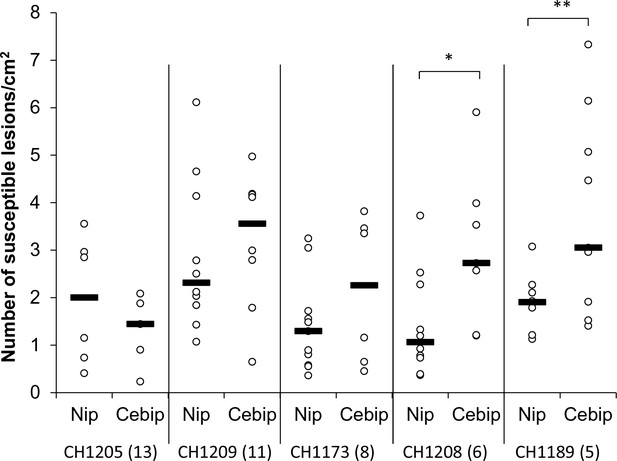
A large complement of effector is no required on immune-deficient plants.
The requirement for Avr-Effectors depends on plant pattern-triggered immunity. Yuanyang isolates with variable number of Avr-Effectors were inoculated onto the cebip mutant and the corresponding Nipponbare (Nip) control. The number beside the isolate name corresponds to the number of Avr-Effector genes as evaluated in Figure 2—figure supplement 3. The number of susceptible lesions per surface unit was measured seven days after inoculation. The black bar represents the median of all data from three replicates. For each isolate, the significance of the difference between cebip and Nipponbare was tested using a T-test (*p<0.05; **p<0.01).
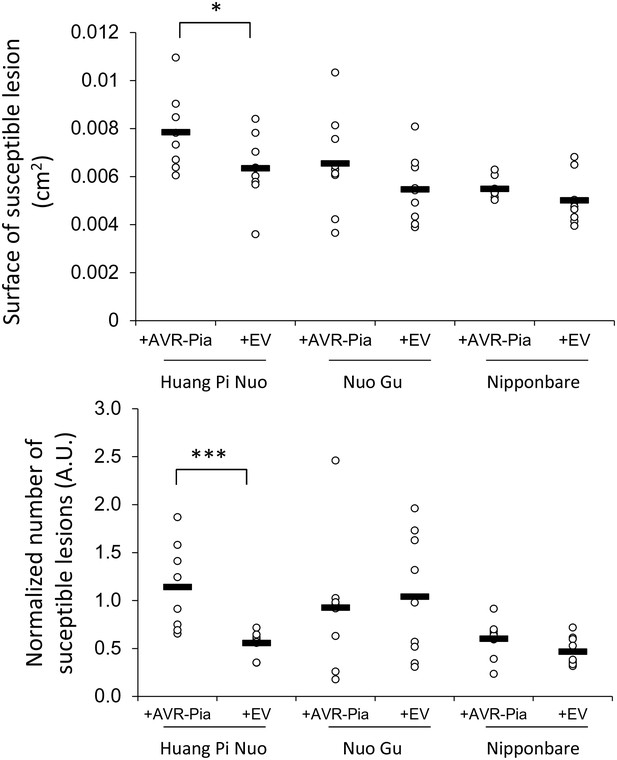
Impact of the Avr-Pia gene on the virulence of M. oryzae on Yuanyang japonica rice varieties.
Three independent transgenic isolates expressing the Avr-Pia gene under its native promoter in the Guy11 background (+Avr-Pia) or three transgenic containing an empty vector (+EV) were inoculated onto the two major japonica varieties grown in Yuanyang (Huang Pi Nuo and Nuo Gu) and two other japonica variety Nipponbare. On Huang Pi Nuo, Avr-Pia isolates are significantly different from isolates containing an empty vector (ANOVA followed by T-test; *p<0.05; ***p<0.001) for two parameters of virulence, the percentage of susceptible lesions (lower panel) and the surface of individual lesions (upper panel). Bars represent average values based on three biological replicates for each of the three Avr-Pia and control strains.
-
Figure 5—source data 1
The data relates to Figure 5.
Strains 1 to 3 are independent GY11 transformants with either AVR-Pia transgene or empty vector (EV). Each strain was replicated three times. HPN= Hunag Pi Nuo, NG= Nu Gu, NB= Nipponbare Spreadsheet 5A and 5B are data for surface of susceptible lesions and susceptible lesions over total lesions.
- https://doi.org/10.7554/eLife.19377.030
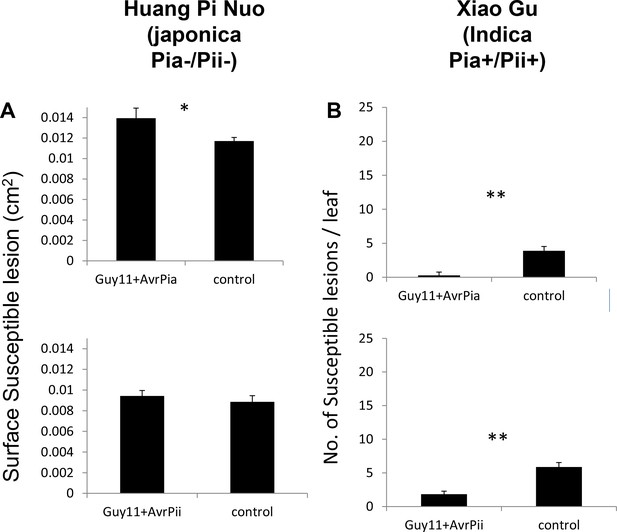
The Avr-Pia but not Avr-Pii gene affects virulence.
(A) Evaluation of the effect of Avr-Effectors on the virulence on the japonica rice Huang Pi Nuo (not containing the corresponding Pi genes) and (B) on indica rice Xiao Gu (containing the corresponding Pi genes). For Xiao Gu, the number of susceptible lesions is given since this genotype was resistant to the isolates containing any of the three AVR gene tested. For Huang Pi Nuo, the GUY11+AVR isolates were virulent and the surface of individual lesions is provided as an indication of virulence.





