Characterization of human translesion DNA synthesis across a UV-induced DNA lesion
Figures
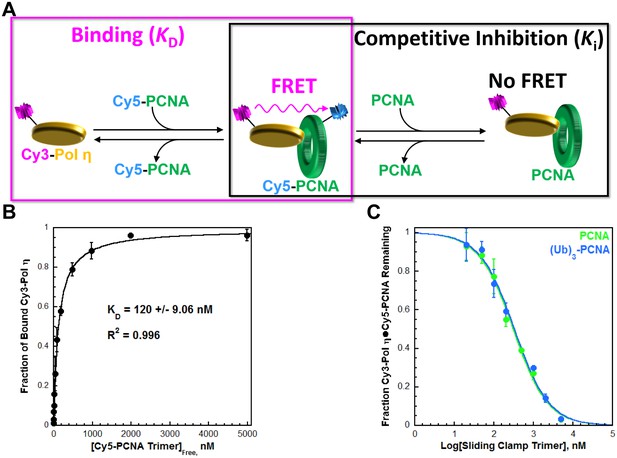
Characterizing the interaction between pol η and PCNA.
Human pol η contains a UBD and three PCNA-binding domains that each resemble those from pol δ (Figure 1—figure supplement 1). We utilized FRET (Figure 1—figure supplement 2) to characterize the interaction between full-length, human pol η and PCNA. (A) Schematic representation of the equilibrium binding assay (left panel) to measure the binding affinity of Cy3-pol η for Cy5-PCNA (Figure 1B) and the competitive inhibition assay (right panel) to measure the binding affinity of an unlabeled PCNA for Cy3-pol η (Figure 1C). (B) Cy3-pol η was kept constant, titrated with Cy5-PCNA, and the fraction of bound Cy3-Pol η was measured. Data was plotted as a function of free Cy5-PCNA concentration and each point represents the average ± SD of 3 independent experiments. Fitting to a one-site binding model, yields a KD of 120 ± 9.06 nM. (C) The Cy3-Pol η•Cy5-PCNA complex was pre-assembled, titrated with an unlabeled PCNA, and the fraction of the Cy3-Pol η•Cy5-PCNA complex remaining was measured. Data was plotted versus the log concentration (in nM) of the respective unlabeled PCNA and each point represents the average ± SD of at least 3 independent experiments. IC50 values for PCNA (green) and (Ub)3-PCNA (blue) were obtained by fitting to a dose-response inhibition model. As a control, this assay was repeated with pol η (Figure 1—figure supplement 3). Ki values were calculated from the IC50 for each competitor and reported in Table 1. (Ub)3-PCNA contains a single ubiquitin moiety on K164 of each monomer within a homotrimeric clamp ring (Figure 1—figure supplement 4).
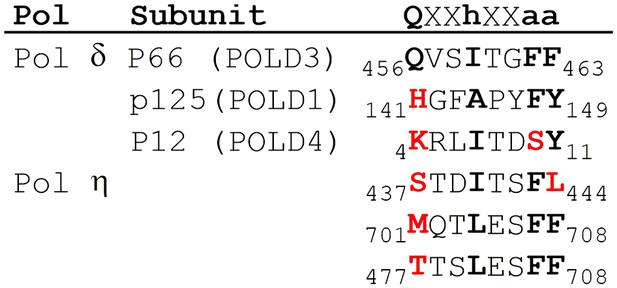
PCNA-binding domains of human pol δ and η.
Sequence alignment of PCNA-interacting peptide (PIP) boxes from human pols δ (Bruning and Shamoo, 2004) and η (Masuda et al., 2015). The generic amino acid sequence of a canonical PCNA-interacting peptide (PIP) box is shown at the top where ‘X’ represents any amino acid, ‘h’ is a hydrophobic residue (usually L, I, or M), and ‘a’ is an aromatic residue (usually F or Y). The canonical PIP-box elements are shown in bold. Non-canonical PIP boxes contain deviations from the conserved residue (shown in bold red).
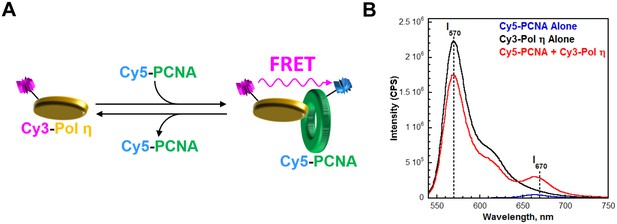
Monitoring the interaction between Pol η and PCNA through FRET.
(A) Schematic representation of the experiment to monitor FRET under equilibrium conditions. Pol η was N-terminally labeled with Cy3 (Cy3-Pol η). Compared to the unlabeled protein, Cy3-Pol η bound to PCNA with similar affinity (Figure 1—figure supplement 3) and retained more than 80% of the DNA synthesis activity (see Materials and methods), demonstrating that Cy3 does not compromise pol η activities. Human PCNA was site-specifically labeled with Cy5 (Cy5-PCNA) at amino acid residue 107, as previously described (Hedglin et al., 2013b). This amino acid residue is located on the ‘back’ face of the PCNA homotrimer, opposite that which interacts with pol η. Interaction between the Cy3-pol η and the Cy5-PCNA was assessed by exciting the Cy3-Pol η with a 514 nm light source and monitoring the fluorescence emission intensities at 570 nm (Cy3 donor fluorescence emission max, I570) and 670 nm (Cy5 acceptor fluorescence emission max, I670). Cy5-PCNA can be excited through FRET from Cy3-Pol η only when the two dyes are in close proximity (<∼10 nm). This is indicated by an increase in I670 (B) Fluorescence emission spectra (540–750 nm) recorded after exciting with a 514 nm light source. The intensities at 670 nm (I670) and 570 nm (I570) are indicated. In the presence of both proteins (200 nM Cy3-pol η, 20 nM Cy5-PCNA), a dramatic enhancement of I670 was observed compared to Cy5-PCNA alone (20 nM). Furthermore, a concomitant decrease in I570 was observed compared to Cy3-pol η alone (200 nM). Such behavior is diagnostic of a FRET signal and indicates a protein-protein interaction between Cy3-pol η and Cy5-PCNA.
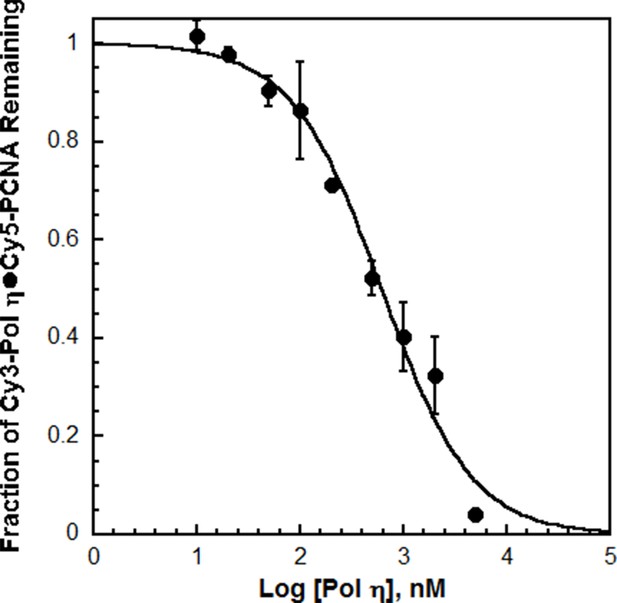
Cy3 does not compromise the affinity of pol η for PCNA.
The competitive inhibition assay (Figure 1C) was repeated with pol η as the unlabeled competitor. The data was plotted versus the log concentration of the unlabeled pol η. Each point represents the average ± SD of 2 independent experiments. The data was fit to a dose-response inhibition model to obtain an IC50 value that was utilized to calculate Ki for pol η. Such analysis yielded a Ki of 112 ± 15.5 nM for pol η, in excellent agreement with the KD for Cy3-pol η (120.1 ± 9.06 nM). This demonstrates that Cy3 does not compromise the ability of pol η to bind PCNA and confirms the validity of the experimental approach.
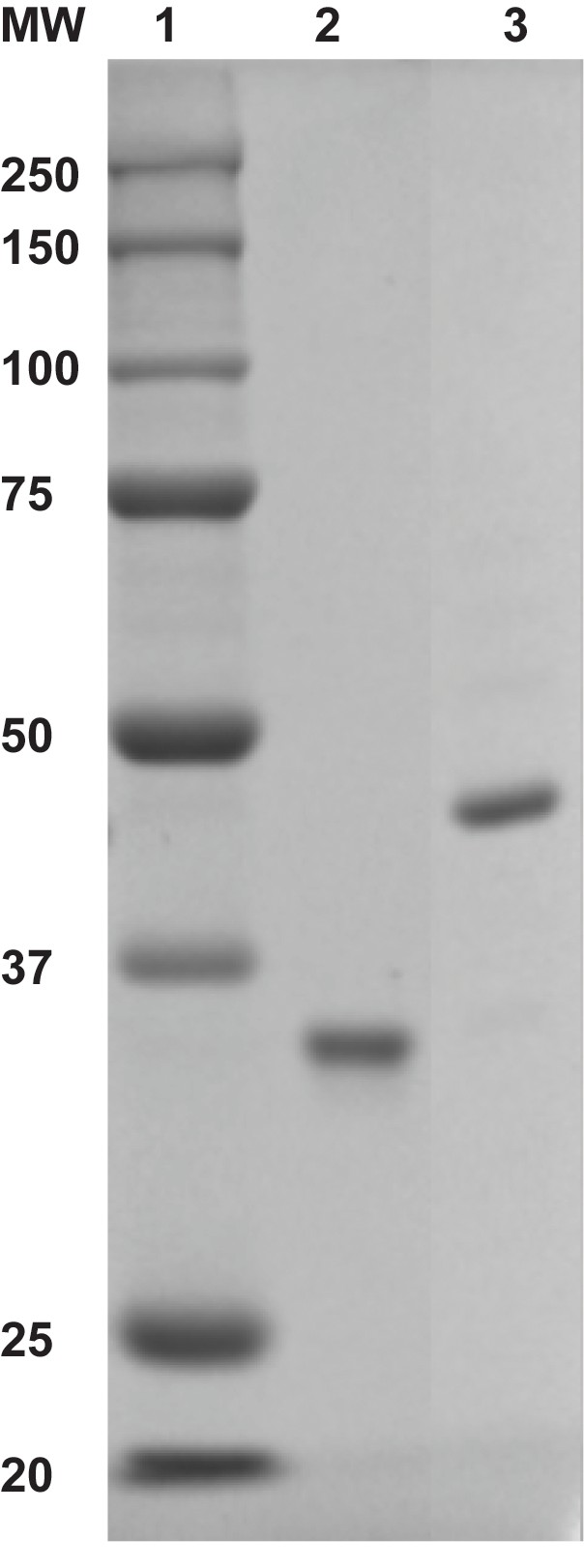
Monoubiquitinated PCNA contains a single ubiquitin moiety on K164 of each monomer within a homotrimeric clamp ring.
(Ub)3-PCNA (10 pmol monomer, Lane 3) was analyzed by coomassie-blue staining after denaturing 12% SDS-PAGE. Lane 1 contains a molecular weight (MW) marker (sizes in kD are indicated on left) and Lane 2 contains wild-type PCNA (10 pmol monomer). The molecular weight of PCNA (Lane 2) is increased approximately 8.5 kDa by the attachment of a single ubiquitin moiety.
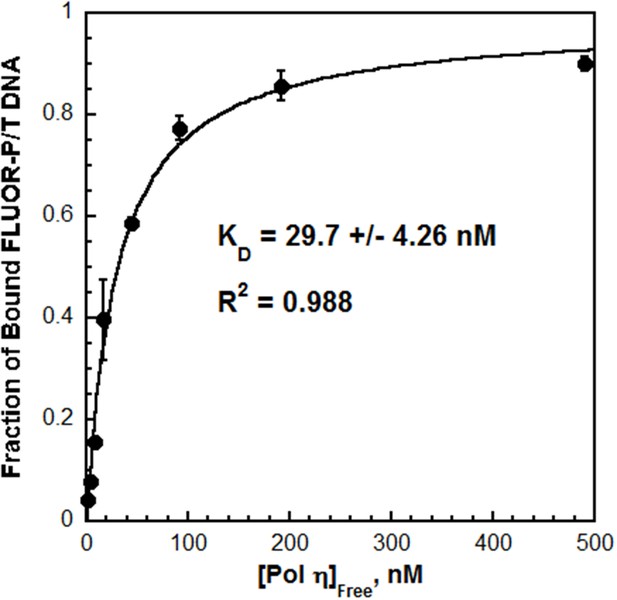
The affinity of Pol η for P/T DNA.
Fluorescein-labeled P/T DNA (FLUOR-P/T, Figure 2—figure supplement 1) was titrated with pol η and the fraction of bound DNA was measured by fluorescence anisotropy. Data was plotted versus the concentration free pol η and each point represents the average ± SD of at least 3 independent experiments. Fitting to a one-site binding model yields a KD value of 29.7 ± 4.26 nM (Table 1). This value is more than 18.1-fold less than the KD estimated (~537 nM) for pol δ on the same DNA substrate (Figure 2—figure supplement 2).
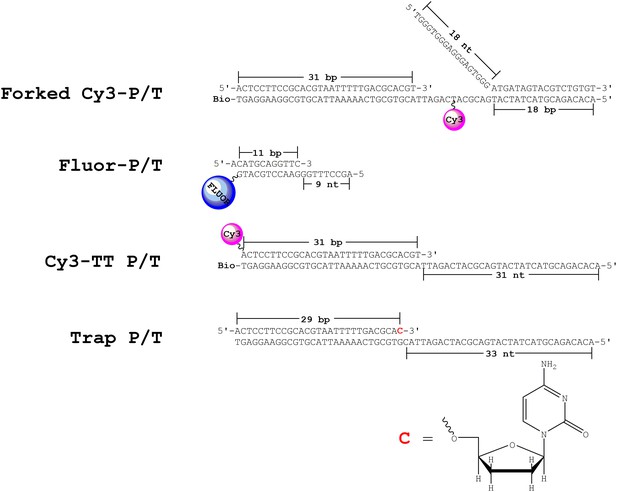
DNA substrates.
The forked Cy3-P/T and Cy3-TT P/T DNA substrates contain Cy3 donor labels and were utilized for FRET-based studies. The FLUOR-P/T DNA substrate contains a 3’-terminal fluorescein label and was utilized for anisotropy measurements. The Trap P/T substrate contains a 3’-dideoxy-terminated primer and was utilized as a passive trap to monitor primer extension under single turnover conditions. denotes a dideoxy-terminating cytosine nucleotide and the structure is shown in the bottom panel.
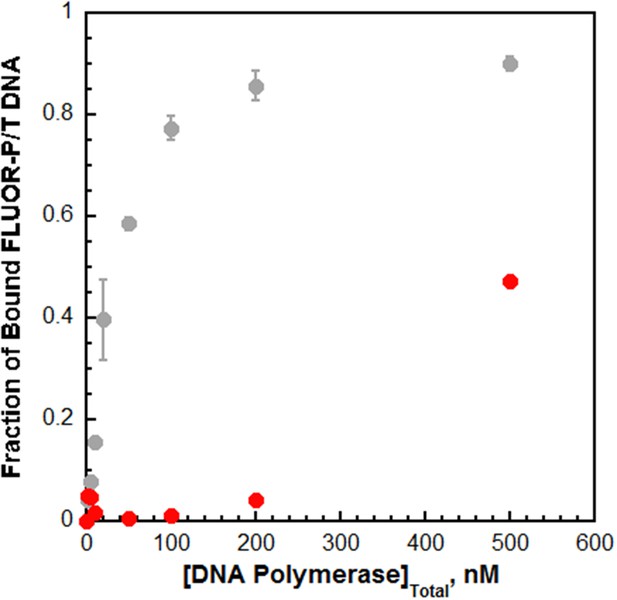
Pol δ binds much weaker to P/T DNA compared to pol η.
The interaction of pol δ with P/T DNA was monitored by fluorescence anisotropy as depicted in Figure 2 Each point is a single measurement for pol δ (). For comparison, the data is overlaid on the data from Figure 2 for pol η (shown in grey). Fractional saturation values for pol δ were estimated from the range observed for the pol η binding curve. The fractional saturation of FLUOR-P/T DNA remained unchanged up to 200 nM pol δ and a significant change was not observed until 500 nM pol δ was added, suggesting that the DNA binding affinity of pol δ is dramatically lower (>10 fold) than that for pol η. In order to maintain physiological ionic strength (200 mM), higher concentrations of pol δ could not be added and, hence, a complete binding curve could not be obtained. Based on the range for the pol η binding curve and the experimental value measured at 500 nM pol δ, a KD of 537 nM is estimated for pol δ, indicating that pol η binds much tighter to P/T DNA (~18.1 fold) than pol δ does.
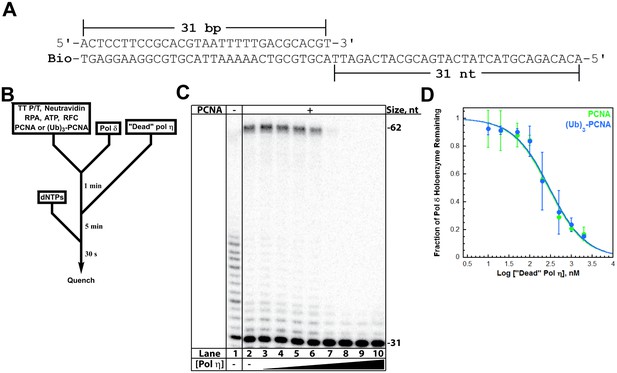
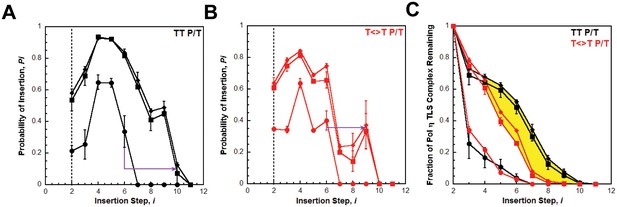
Polymerase exchange at a blocked P/T junction.
(A) Sequence of the TT P/T DNA substrate that mimics a nascent P/T junction on the lagging strand. The size of the double-stranded DNA region agrees with the size of an initiating P/T and the requirements for assembly of a pol δ holoenzyme. The biotin tag was pre-bound to Neutravidin, preventing PCNA from sliding off the 5’ end of the primer. The ssDNA is consistent with the footprint of a single RPA molecule and was pre-bound with excess RPA. The primer is labeled at the 5’ terminus with 32P (Hedglin et al., 2016). (B) Schematic representation of the experiment to monitor pol exchange. The experiment for pol δ alone was performed identically except for the omission of PCNA, RFC, and ‘dead’ pol η. (C) 12% denaturing sequencing gel of the primer extension products. The size of the substrate and full-length product are indicated on the right. The full-length product is only generated in the presence of a PCNA (compare lane 1 to lanes 2–10). Thus, production of the full-length product indicates the fraction of pol δ holoenzymes that withstand the influx of ‘dead’ pol η and, hence, the extent of pol exchange. (D) The fraction of pol δ holoenzyme remaining was plotted as a function of the log concentration of ‘dead’ pol η. The data for pol δ holoenzymes assembled with either PCNA (green) or (Ub)3-PCNA (blue) is shown and each point represents the average ± SD of 3 independent experiments. IC50 values were obtained by fitting the data to a dose-response inhibition model and Ki values (Table 1) were calculated from the IC50 values. Ki values of 31.7 ± 5.12 nM and 33.5 ± 5.04 nM (Table 1) were calculated for pol δ holoenzymes assembled with PCNA and (Ub)3-PCNA, respectively. These equivalent values exactly agree with the KD for pol η binding to P/T DNA (Figure 2) and the KD for pol η holoenzyme formation (Figure 3—figure supplement 1), suggesting that pol η binding to the P/T junction is competing with pol δ binding to PCNA encircling the P/T junction. Repeating this assay with Cy5-PCNA and Cy3-labeled TT P/T DNA and monitoring FRET instead of DNA synthesis (Figure 3—figure supplement 2) verified that PCNA does not slide off the unblocked end of the DNA substrate template during the competition.
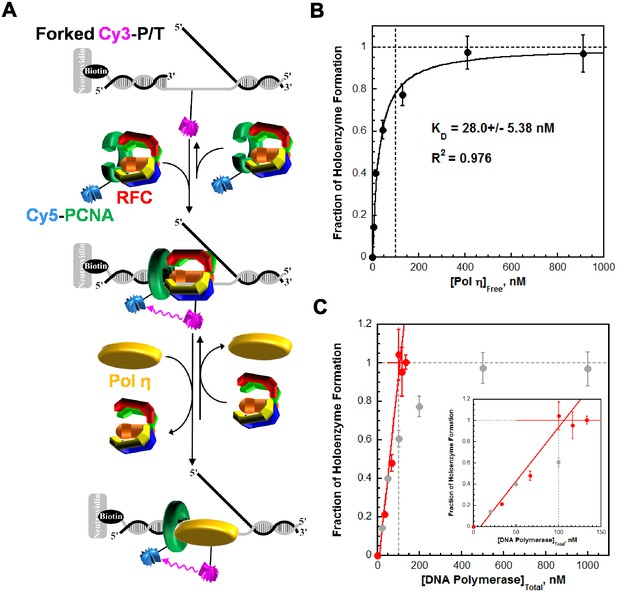
Differential binding activities drive assembly of the pol η and pol δ holoenzymes.
Assembly of a pol η holoenzyme was monitored via FRET by slight modifications to a published protocol (Hedglin et al., 2013b). (A) Schematic representation of the experiment depicted in B to monitor FRET under equilibrium conditions. A forked P/T DNA substrate (forked Cy3-P/T DNA) in agreement with the minimal requirements for assembly of human RFC and PCNA onto DNA was labeled with an internal Cy3 dye. This substrate also carries a 3′-biotin label. Together with the flap, the 3′-biotin label in complex with Neutravidin prevents loaded clamp from sliding off the DNA. Forked Cy3-P/T DNA (100 nM) was pre-incubated with Cy5-PCNA (100 nM), RFC (100 nM), and ATP (1 mM). Under these stoichiometric conditions, RFC loads all Cy5-PCNA onto Cy3-DNA and, in the absence of polymerase, dissociates back into solution taking all loaded PCNA with it. After the initial loading event, the clamp loading-unloading pathway reaches equilibrium where the rate constants for PCNA loading and unloading are equal and a net change in the FRET signal is no longer observed. At equilibrium, PCNA loading is highly-favored due to a much faster rate constant for PCNA loading compared to PCNA unloading (Hedglin et al., 2013b). During holoenzyme formation, an incoming polymerase captures loaded PCNA from DNA-bound RFC, stabilizing the sliding clamp on DNA and increasing the fraction of bound forked Cy3-P/T DNA•Cy5-PCNA. (B) The fractional saturation increased with free Pol η concentration and displayed a hyperbolic behavior indicative of an equilibrium binding curve. The dashed lines indicate the concentrations of Cy3-P/T DNA, Cy5-PCNA, and RFC (100 nM each, X = 100 nM) and 100% holoenzyme formation (y = 1.0). The data fit best to a one-site binding model yielding a KD of 28.0 ± 5.38 nM for the forked Cy3-P/T DNA•Cy5-PCNA•Pol η complex. Such behavior is in stark contrast to that observed for pol δ in a recent report from our lab, shown in C. (C) Fractional saturation of the P/T DNA•PCNA complex as a function of the total concentration of DNA polymerase. Shown in grey is the data from B for pol η. Shown in red is the data for pol δ from reference (Hedglin et al., 2013b). In contrast to pol η, pol δ stabilized a stoichiometric amount of PCNA on DNA under the same conditions and, hence, displayed a linear increase. The line plateaued when the concentrations of total pol δ, PCNA, and RFC were equivalent (100 nM of each), and flat-lined thereafter (Hedglin et al., 2013b). This indicates that the total concentration of the DNA•PCNA complex (100 nM) is much greater (>10 fold) than the KD of pol δ for the DNA•PCNA complex (Goodrich and Kugel, 2007). This sets an upper limit of 10 nM for the KD of pol δ for PCNA loaded onto DNA, in excellent agreement with the value reported in an independent study (7.1 ± 1.0 nM) (Zhou et al., 2012).
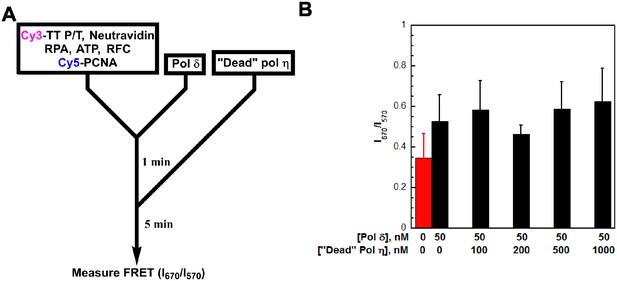
The resident PCNA remains at the blocked P/T junction during the exchange of pol δ for pol η.
(A) Schematic representation of the experiment depicted in panel B to monitor the FRET between Cy5-PCNA and the Cy3-TT P/T DNA substrate. The TT P/T DNA substrate now contains a 5’ Cy3 label on the primer (Cy3-TT P/T DNA, Figure 2—figure supplement 1). The pol δ holoenzyme was pre-assembled with Cy5-PCNA on the Cy3-TT P/T DNA substrate as described in Figure 3 Catalytically-inactive (‘dead’) pol η (0–1 μM) was added and the FRET (I670/I670) was measured after a 5 min incubation (B) Data from experiments described in panel A plotted as a function of ‘dead’ pol η concentration. Each column represents the average ± SD of 3 independent experiments. As a control, the FRET was measured in the absence of pol δ (). Under these conditions, pol δ (50 nM) stabilizes Cy5-PCNA (50 nM) on the Cy3-TT P/T DNA substrate (10 nM), maximizing the FRET signal. During the exchange of pols, if Cy5-PCNA slides off the unblocked end of the template that lacks biotin/neutravidin, the FRET signal will decrease as the concentration of ‘dead’ pol η is increased. However, the FRET signal remains constant up to 1 μM ‘dead’ pol η where essentially all of pol δ holoenzyme residing at the P/T junction has been exchanged (Figure 3). This demonstrates that PCNA does not slide off the TT P/T DNA substrate during the exchange of pol δ for pol η.
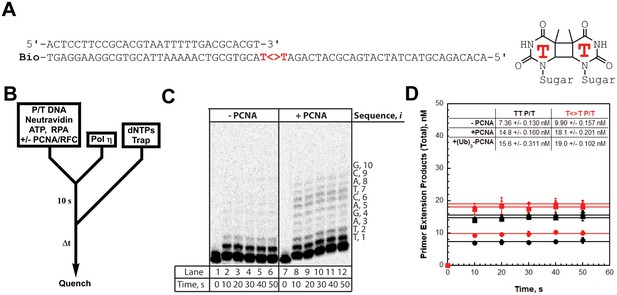
DNA synthesis by pol η.
(A) Sequence of the T<>T P/T DNA substrate. This substrate is identical to the undamaged DNA substrate (Figure 3A) except the native TT sequence has been replaced with a TT-CPD (denoted , shown on right). (B) Schematic representation of the experiment performed to monitor primer extension by pol η under single turnover conditions. The experiment for pol η alone was performed identically except for the omission of PCNA and RFC. (C) 16% denaturing sequencing gel of the primer extension products for pol η alone (Lanes 1–6) and a pol η holoenzyme assembled with PCNA (lanes 7–12). The sequence of the template for each dNTP insertion step (i) is indicated on the right up to i = 10. (D) Quantification of the primer extension products observed in the absence of a PCNA (●), in the presence of PCNA (▪), and in the presence of (Ub)3-PCNA (♦). Results for undamaged (TT P/T) and damaged () DNA substrates are shown. The data is plotted versus time and each data point represents the average ± SD of 3 independent experiments. Data points after t = 10 s were fit to a flat line where the y-intercept reflects the amplitude. Values are reported in the inset. In contrast to that observed for pol η on the undamaged (TT P/T) DNA substrate, DNA synthesis by pol δ was only observed in the presence of PCNA (Figure 4—figure supplement 1).
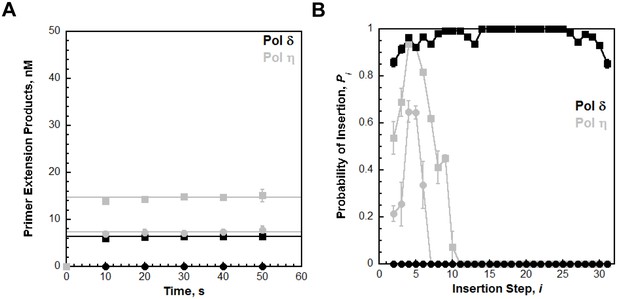
Processive DNA synthesis by pol δ.
Primer extension by pol δ was monitored under single turnover conditions on the TT P/T DNA substrate as described in Figure 4 and the probability of insertion (Pi) for each dNTP insertion step (i) was calculated. (A) Quantification of the primer extension products observed in the absence (●) and presence of PCNA (▪). The results are plotted versus time and each data point represents the average ± SD of 3 independent experiments. Data points after t = 10 s were fit to a flat line where the y-intercept reflects the amplitude. For comparison, the data was overlaid on the data obtained for pol η (shown in grey) on the same substrate (Figure 4D) (C) The probability of insertion (Pi) for each step (i) beyond the first insertion observed in the absence (●) and presence of PCNA (▪). The results are plotted versus the insertion step and each data point represents the average ± SD of at least 3 independent experiments. For comparison, the data is overlaid on the data obtained for pol η (shown in grey) on the same substrate (Figure 4A). In contrast to pol η, DNA synthesis by pol δ is only observed in the presence of PCNA. Hence, the measured Pi values only reflect dNTP insertion by pol δ holoenzymes. From i = 15 to i = 25 for pol δ, Pi plateaus and remains constant in the presence of PCNA. Within this range, Pi is 0.999 ± 1.96 × 10−4 for pol δ holoenzymes, in excellent agreement with a previous report (Hedglin et al., 2016).
Processive DNA synthesis by pol η.
Probability of insertion (Pi) for each dNTP insertion step (i) beyond the first was calculated for the experiments depicted in Figure 4. Results obtained in the absence of PCNA (●), in the presence of PCNA (▪), and in the presence of (Ub)3-PCNA (♦) are plotted versus the insertion step up to i = 12 and each data point represents the average ± SD of at least 3 independent experiments. dNTP insertion beyond i = 10 was not observed for pol η. (A –B) Processivity of pol η on undamaged (panel A, TT P/T) and damaged (panel B, ) P/T DNA substrates. The dashed line denotes the population of pol η that inserts a dNTP across from the 5’ T of a given template prior to dissociation, that is the pol η TLS complex. The arrows denote the increase in the extent of continuous DNA synthesis observed in the presence of a PCNA. (C) Dissociation of the pol η TLS complex. The fraction of the Pol η TLS complex (y) remaining at each insertion step, i, was determined for the undamaged (TT P/T) and damaged () P/T DNA substrates using the Pi values reported in panels A and B. Results obtained in the absence of PCNA (●), in the presence of PCNA (▪), and in the presence (Ub)3-PCNA (♦) are plotted versus the insertion step, i. The divergence observed in the presence of a PCNA for the damaged and undamaged substrates is highlighted in yellow.
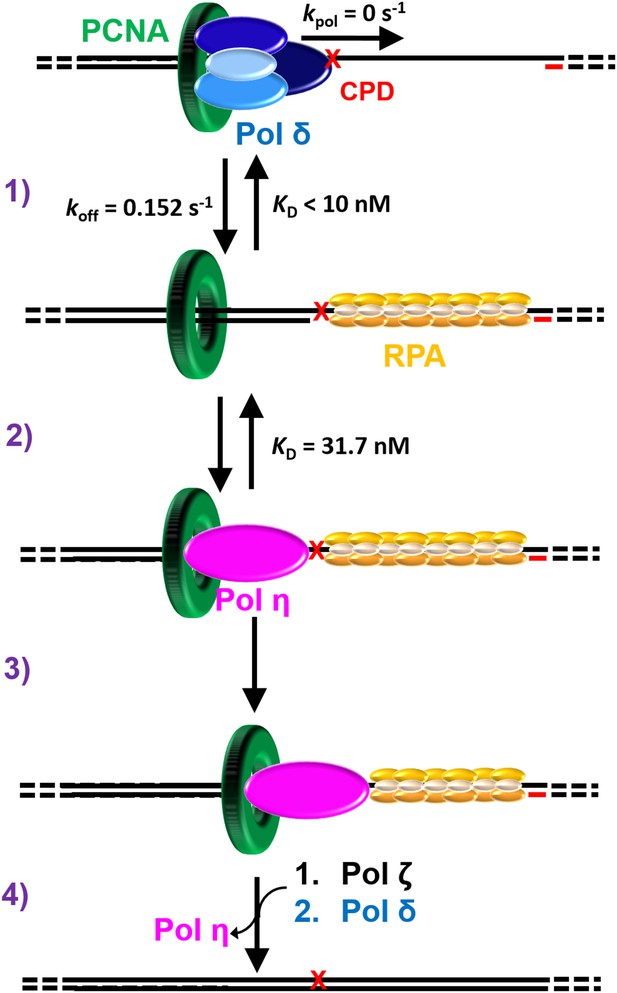
Polymerase exchange during TLS on the lagging strand.
While replicating a lagging strand template, pol δ maintains a loose association with PCNA while inserting dNTPs very fast. (1) Upon encountering a DNA lesion () it cannot accommodate, such as UV-induced CPD, pol δ rapidly dissociates from DNA, leaving PCNA behind. Pol δ may re-bind to the resident PCNA but pol δ-mediated DNA synthesis cannot resume until the lesion is replicated by one or more TLS pols. (2) The binding affinity of pol η for P/T DNA drives the passive exchange of pols at a blocked P/T junction. (3) Once localized to the blocked P/T junction, the resident PCNA stabilizes pol η on the DNA, increasing the efficiency of the ensuing TLS. (4) After replicating the CPD, dissociation of pol η is promoted downstream of the damage. This provides an efficient and generic pathway for pol η to depart a damaged template and, hence, limits the extent of error-prone DNA synthesis by pol η downstream of a CPD. In the event that pol η-mediated TLS across and beyond a CPD lesion terminates with a mismatched P/T junction, another TLS pol, such as pol ζ, may access the P/T junction upon pol η’s departure and faithfully extend the mismatch, allowing DNA synthesis by pol δ to resume.
Tables
Binding affinities (KD or Ki) calculated from equilibrium assays carried out in the present study.
| Binding assays | Substrate | Ligand | KD, nM | Figure* |
|---|---|---|---|---|
| Cy3-Pol η | Cy5-PCNA | 120 ± 9.06 | 1B | |
| FLUOR-P/T DNA | Pol η | 29.7 ± 4.26 | 2 | |
| Pol δ | ~537 | 2FS2 | ||
| Forked Cy3-P/T DNA•Cy5-PCNA | Pol η | 28.0 ± 5.38 | 3FS1 | |
| Pol δ | < 10.0 | 3FS1 |
| Competitive inhibition assays | Complex | Competitor | Ki, nM | Figure* |
|---|---|---|---|---|
| Cy3-Pol η•Cy5-PCNA | PCNA | 111 ± 7.78 | 1C | |
| (Ub)3-PCNA | 119 ± 10.5 | 1C | ||
| Pol η | 112 ± 15.5 | 1FS3 | ||
| P/T DNA•PCNA•Pol δ | 'Dead' pol η | 31.7 ± 5.12 | 3 | |
| P/T DNA•(Ub)3-PCNA•Pol δ | 'Dead' pol η | 33.5 ± 5.04 | 3 |
-
*Figure 2—figure supplement 2, Figure 3—figure supplement 1, and Figure 1—figure supplement 3 are abbreviated as 2FS2, 3FS1, and 1FS3, respectively.





