Calcium-mediated actin reset (CaAR) mediates acute cell adaptations
Figures
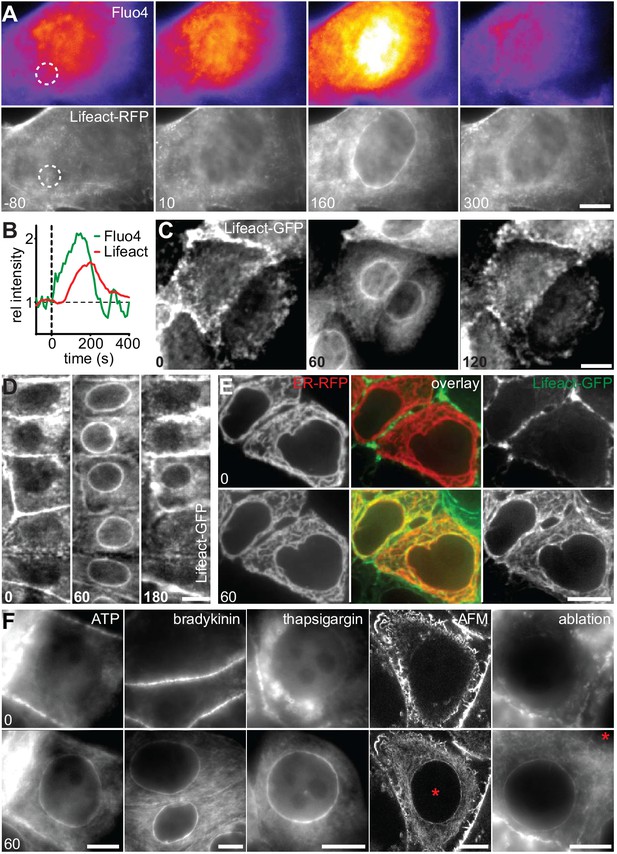
A calcium-mediated actin reset in mammalian cells.
(A, B) MDCK cells labeled with Lifeact-mCherry (Lifeact-RFP) and Fluo4 were exposed to 10 dyn/cm2 shear flow. Regions used for intensity plots in (B) are indicated in (A). (C) MDCK cells expressing Lifeact-GFP were stimulated with 1 µM ionomycin. (D–F) MCF-7 cells expressing Lifeact-GFP (and ER-RFP in (E)) were exposed to 1 µM ionomycin (D, E), or to 50 µM ATP, 1 µM bradykinin or 1 µM thapsigargin or locally stressed with an AFM probe (pointy tips) or by laser ablation (F) (asterisks at position of stimulus). Times in sec. Scale bars: 10 µm.
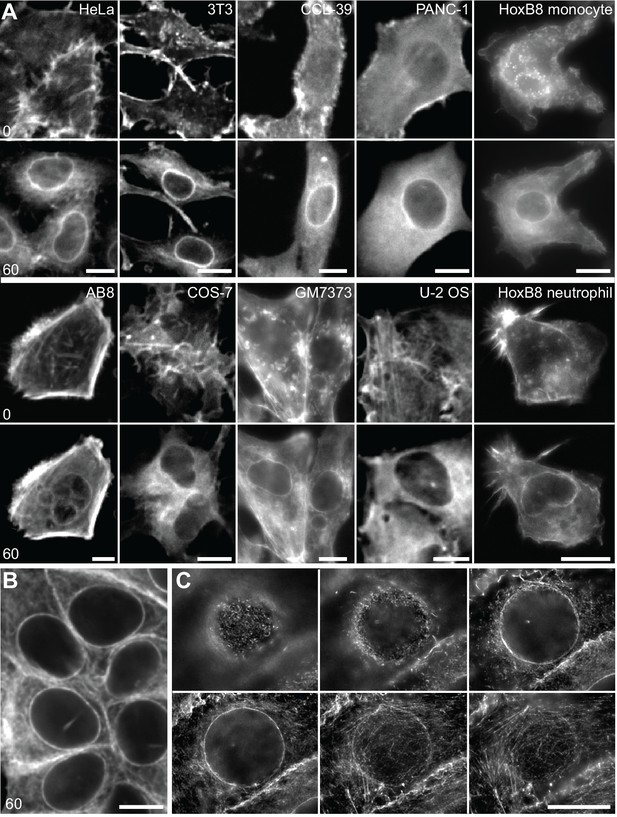
A calcium-mediated actin reset in mammalian cells.
(A) Treatment of indicated cell types expressing Lifeact-GFP with 1 µM ionomycin. (B, C) MCF-7 cells were treated with 1 µM ionomycin for 1 min, fixed and stained with phalloidin-Alexa647. Images in (C) represent a series of z-planes acquired at 0.5 µm distance. Times in sec. Scale bars 10 µm.
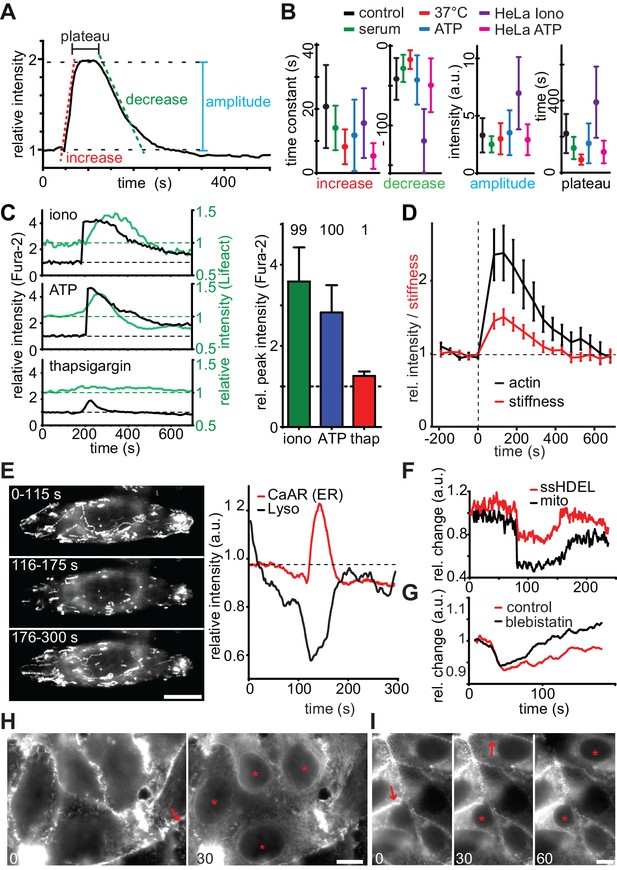
Quantitative analysis of CaAR.
(A, B) Quantification of indicated parameters during CaAR. MCF-7 and HeLa cells expressing Lifeact-GFP were incubated at RT or 37°C and with serum or serum-free HBSS buffer. Cells were then treated with either 330 nM ionomycin or 50 µM ATP. Control corresponds to serum starved MCF-7 cells stimulated with 330 nM ionomycin at RT. GFP intensity was measured at the nuclear periphery and analyzed using customized Matlab scripts (Supplementary material). All values are mean ± SD, n > 100 cells. See Table 1 for details. (C) Lifeact-GFP expressing MCF-7 cells were treated with the indicated drugs, ATP: 50 µM, ionomycin: 1 µM, thapsigargin: 1 µM in Ca2+-free medium. Intracellular calcium levels (Fura2) and Lifeact-GFP intensities at the ER were monitored. Peak values: mean ± SD (n > 30). Numbers above bars indicate % cells exhibiting CaAR. (D) Cells treated with 1 µM ionomycin were followed over time by simultaneous fluorescence and atomic force microscopy. The relative Young’s modulus of whole cells was calculated from force-distance curves obtained with 10 µm beads (mean ± SD, n = 24). (E) Freezing of lysosomes labeled with Lysotracker Red in HeLa cells undergoing CaAR. Images correspond to the maximum projection of indicated periods in a time series. (F) Freezing of organelle motion during CaAR in MCF-7 cells. ER or mitochondria were fluorescently labeled with ss-RFP-KDEL or mitotracker Red, respectively. (G) Change in lysosome motility for control and blebbistatin-treated (50 µM) HeLa cells. (H, I) Propagation of CaAR induced by laser ablation in the absence (I) and presence (J) of 50 µM ATP. Arrows: ablation sites, asterisks: cells reacting to stimulus. Times in sec after exposure to the stimulus. Scale bars: 10 µm.
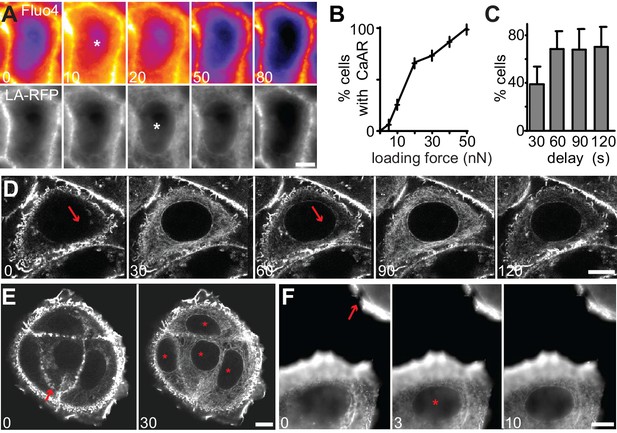
Quantitative analysis of CaAR.
(A) MCF-7 cells expressing Lifeact-mCherry (LA-RFP) were labeled with Fluo4 and stimulated by laser ablation. Asterisks: first frame in which Fluo4 or CaAR response can be detected. (B) Incidence of CaAR (in %) induced in MCF-7 cells by the application of different loading forces with the AFM probe (pointy tip). n = 10 cells per data point. (C) Efficiency of repeated cell stimulation with varying delays (mean ± SD, n = 7). (D) Repeated stimulation of cells with pointy AFM tip. (E, F) CaAR induction and propagation in MCF-7 cells following stimulation by AFM (E) or laser ablation (F). Arrow: ablation site / AFM contact; asterisks: cells reacting to stimulus. Times in sec. Scale bars: 10 µm.
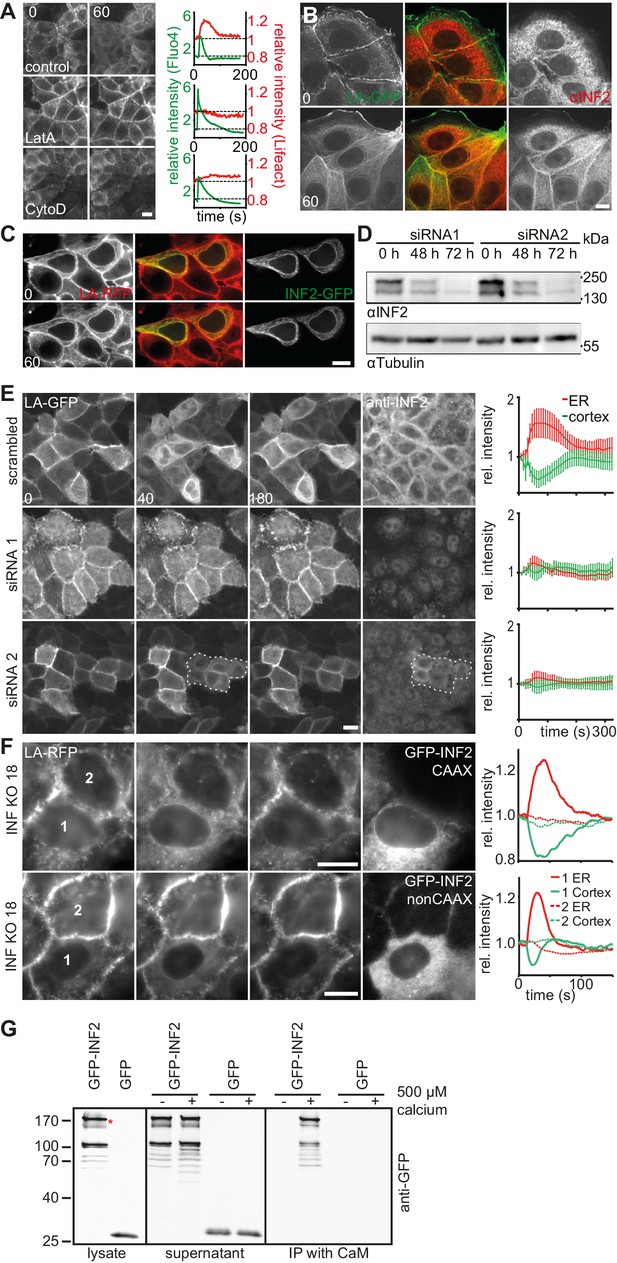
CaAR is driven by INF2-mediated actin polymerization.
(A) MCF-7 cells expressing Lifeact-mCherry were treated with ionomycin and the indicated drugs (LatA: 400 nM latrunculin A, CytoD: 1 µM cytochalasin D). Plots show representative intensity profiles for Lifeact-GFP at the nuclear periphery and for intracellular Ca2+ (Fluo4). (B) MCF-7 cells were fixed at the indicated time points after addition of 1 µM ionomycin and stained with αINF2 antibody. (C) MCF-7 cells expressing Lifeact-mCherry were transfected with a constitutively active GFP-INF2(A149D) construct and imaged before and after addition of 1 µM ionomycin. (D, E) siRNA-mediated knock-down of INF2 in HeLa cells expressing Lifeact-GFP. (D) Western analysis of INF2 after knock-down with two different siRNAs at indicated times. (E) 72 hr after siRNA transfection, HeLa cells were treated with 1 µM ionomycin and monitored by fluorescence microscopy. Cells were fixed, immunostained with anti-INF2 antibody and imaged again at the same positions. Dotted line surrounds residual INF2-positive cells. Plots show Lifeact-GFP intensity at the cortex (green) and the ER (red). Values are mean ± SD, n = 30. (F) Images of HeLa INF2 KO cells stably expressing Lifeact-mCherry (derived from KO clone 18) and transfected with either GFP-INF2-CAAX or GFP-INF2-nonCAAX. Cells were stimulated by laser ablation outside the represented region. In each series one cell with INF2 expression (1) and a control cell without INF2 (2) are labeled. Corresponding intensity plots for ER (red) and cortical (green) regions are shown. Times in sec. Scale bars: 10 µm. (G) Co-precipitation analyses showing that GFP-INF2-CAAX expressed in HEK293 cells specifically interacts with immobilised calmodulin (CaM). Comparison of pull down conditions with and without (1 mM EGTA) 500 µM Ca2+ (CaM activation at plateau).
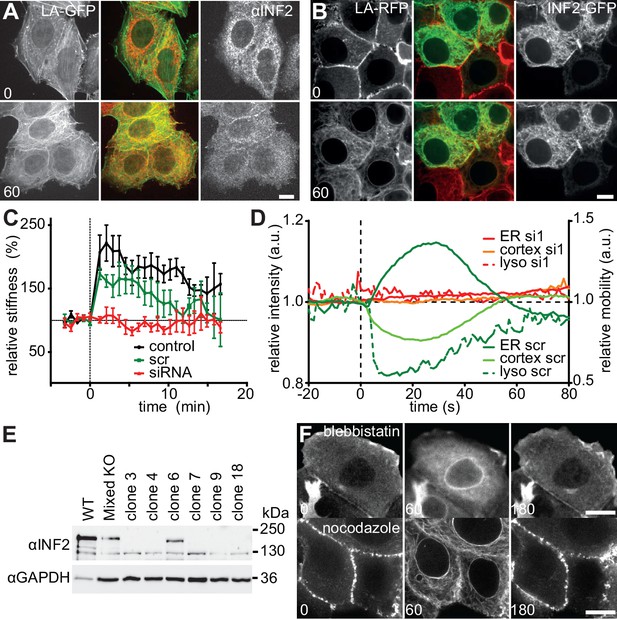
CaAR is driven by INF2-mediated actin polymerization.
(A) HeLa cells expressing Lifeact-GFP were fixed at the indicated time points after addition of 1 µM ionomycin and stained with αINF2 antibody. (B) MCF-7 cells expressing Lifeact-mCherry were transfected with a GFP-INF2 expressing plasmid and induced with 1 µM ionomycin. Times in sec. Scale bars 10 µm. (C) Lifeact-GFP-expressing HeLa cells were transfected with scrambled siRNA or INF2 targeting siRNA1, or left untreated (control) and analyzed by atomic force microscopy. The relative Young’s modulus of whole cells was calculated from force-distance curves obtained with 10 µm beads and plotted as mean ± SD (n > 4). (D) Normalized Lifeact-GFP intensities at the ER and the cortex and mobility of lysosomes labeled with Lysotracker Red of representative HeLa cells stimulated by laser ablation. Cells were observed 72 hr after transfection with scrambled (scr) or INF2 siRNA (si1). (E) Western blot with antiINF2 antibody showing loss of INF2 expression after CRSPR/Cas9-mediated knock out in different isolated clones. (F) CaAR in MCF-7 cells pretreated with 50 µM blebbistatin or 10 µM nocodazole for 20 min. Images were taken at the indicated times after addition of 1 µM ionomycin.
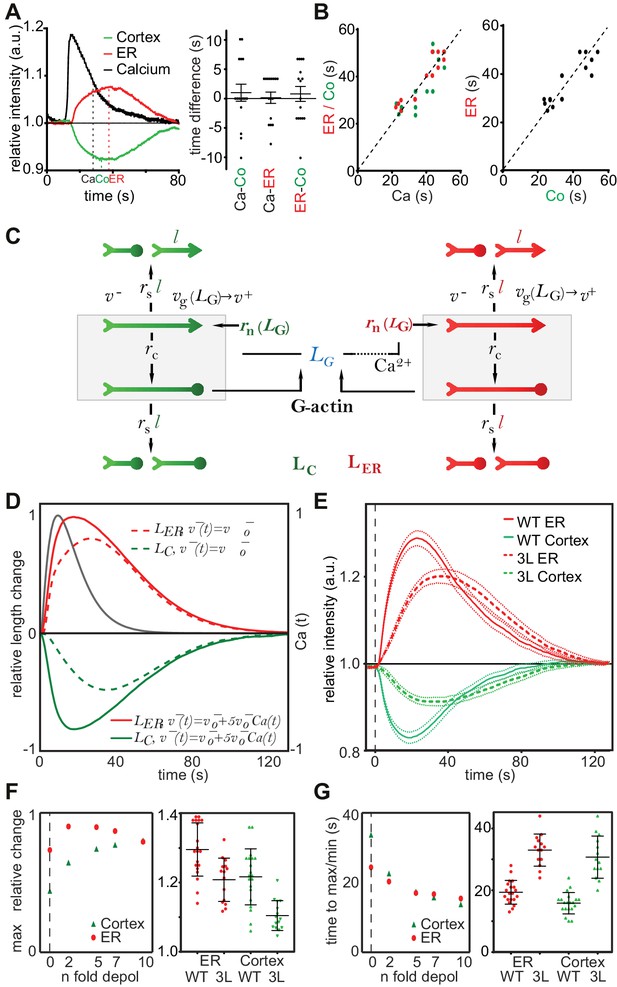
A stochastic model rationalizes CaAR features and kinetics.
(A, B) Correlation analysis of actin and calcium dynamics in MCF-7 cells undergoing CaAR. (A) Temporal shift between half-maximal decay of calcium (Ca), actin maximum at nuclear periphery (ER) and actin minimum at the cell cortex (Co). (B) Correlation between Ca and ER (red, Pearson correlation coefficient, r = 0.924), Ca and Co (green, r = 0.840) and between ER and Co (black, r = 0.880). Dotted lines: y = x. (C) Stochastic model for actin competition. Filaments at the cortex (green) and ER (red) are represented by arrows (head: barbed end, circle: capped end). Total length of cortical (LC) and ER (LER) actin is indicated. Monomer pool is given by LG. Relevant parameters: Rates of severing (rs per length l), capping (rc), nucleation (rn), velocities of elongation (v+) and depolymerization (v-). See Supplementary material for details of the model. (D) Evolution of F-actin concentration at ER (red) and cortex (green) generated by the model using a simple competition scenario (dotted lines) or including calcium-activated depolymerization of actin (full lines). All simulations were run using the indicated Ca2+ curve (black) as input. (E) Average intensity curves of Lifeact-mCherry at ER (red) and cell cortex (green) of HeLa KO cells expressing wildtype or WH2 mutant GFP-INF2-CAAX (3L: L976A, L977A, L986A). Dotted red and green lines indicate SEM. (F) Effect of different depolymerization rates on maximal actin change at cortex (green) and ER (red). Shown are predictions from simulations (left) and experimental data comparing wildtype and 3L-INF2 expressing HeLa cells (data points, mean and SD). (G) As in (F) but showing the effects of varying depolymerization rate on the time until cortical minimum or ER-maximum is reached.
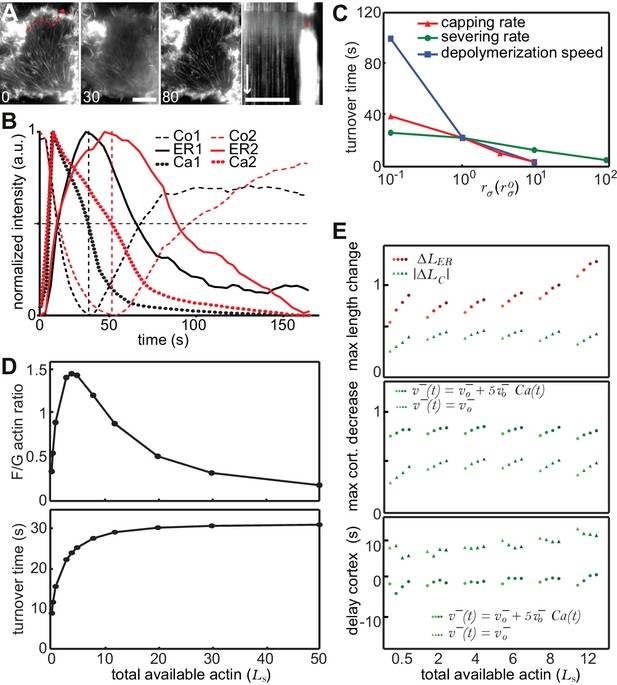
A stochastic model rationalizes CaAR features and kinetics.
(A) TIRF imaging of the basal surface of a Lifeact-GFP expressing MCF-7 cell after laser ablation. The dashed line indicates the position of the kymograph, the asterisk indicates the duration of CaAR. Times in sec, scale bars 10 µm, time arrow 60 s. (B) MCF-7 cells expressing Lifeact-GFP were labeled with Fluo4 and stimulated by laser ablation. Signal intensities for Fluo4 (reflects intracellular calcium levels; dotted curves) and Lifeact-mCherry in the perinuclear area (solid curves) and the cell cortex (dashed curves) were plotted for two cells with shorter (red) and longer (black) calcium peaks. Half maximal calcium levels and corresponding maxima/minima of actin are indicated by dashed vertical lines. (C–E) Impact of changes in the indicated parameters of the stochastic model describing actin reorganization during CaAR. See Materials and methods for details.
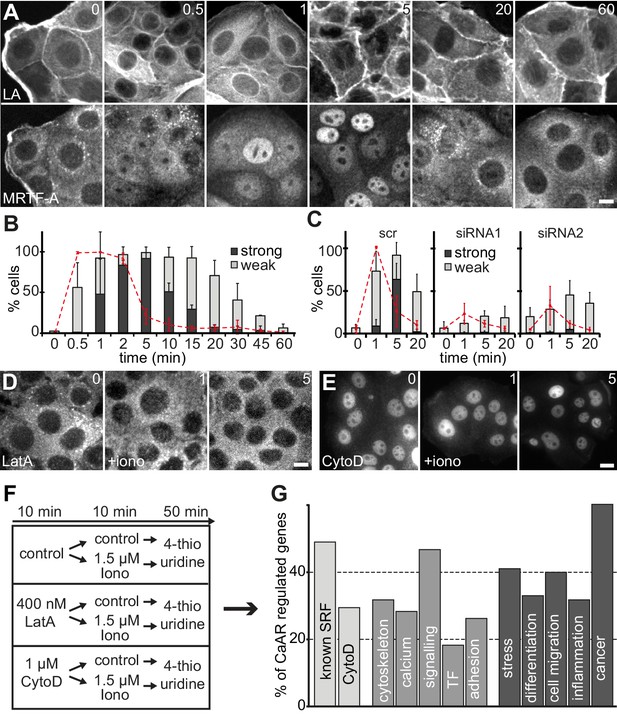
CaAR induces transcription via MRTF-A and SRF.
(A) Immunofluorescence detection of MRTF-A in MCF-7 cells at the indicated times after addition of 1 µM ionomycin. (B) Quantification of MCF-7 cells exhibiting CaAR (red dotted line) and weak (light grey) or strong (dark grey) nuclear MRTF-A accumulation (mean + SD, n = 3 experiments with >100 cells per time point). (C) Quantification of HeLa cells exhibiting CaAR (red dotted line) and weak (light grey) or strong (dark grey) nuclear MRTF-A staining (mean + SD, n = 3 experiments with >100 cells per time point). Graphs show values for cells treated with scrambled siRNA and two INF2-siRNAs. (D, E) Analysis of MRTF-A localization upon stimulation of MCF-7 cells with 1 µM ionomycin (added at t = 0). Cells were pretreated for 10 min with either 400 nM latrunculin A (D) or 1 µM cytochalasin D (E). (F) Protocol for transcriptome analysis. (G) Major functional categories of CaAR-regulated genes, assembled from the manual curation of public databases and literature. Association categories are given for the known SRF regulation (light grey), cellular processes (medium grey) and biological processes (dark grey). Time in min. Scale bars 10 µm.
-
Figure 5—source data 1
CaAR induces transcription via MRTF-A and SRF.
Excel table with 88 genes differentially regulated in MCF-7 cells either treated for 10 min with ionomycin (1.5 µM) or with ionomycin (1.5 µM) and LatA (400 nM). Filled cells indicate inclusion into indicated categories. Orange: regulation through SRF or CytoD-induced MRTF-A translocation. Blue: cellular processes. Green: physiological processes. Total number and % of genes in each category are given at the bottom of the table.
- https://doi.org/10.7554/eLife.19850.021
-
Figure 5—source data 2
Differentially regulated genes.
Excel table (five pages) with results from Affymetrix chip analysis for gene expression after treatment with Ionomycin and actin drugs. See Materials and Methods for details.
- https://doi.org/10.7554/eLife.19850.022
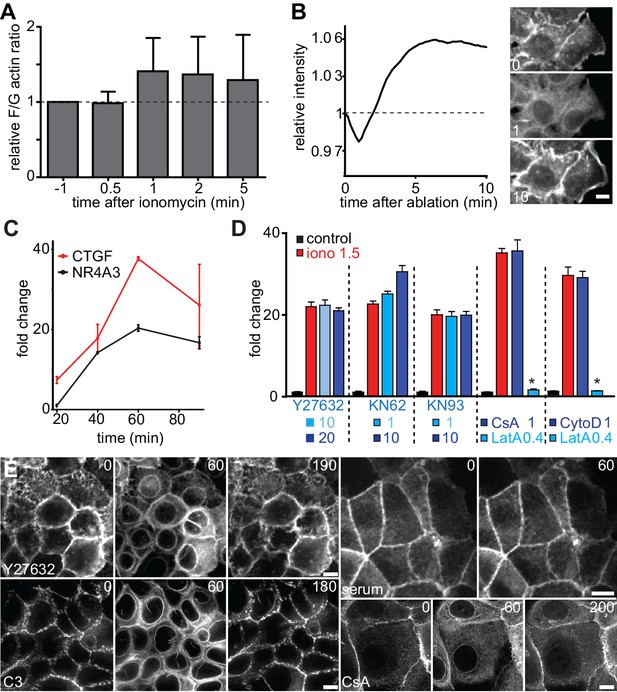
CaAR induces transcription via MRTF-A and SRF.
(A) Ratio of F- to G-actin in MCF-7 cells during CaAR as determined by fractionation and Western blotting (mean ± SD, n = 5). (B) Lifeact-GFP intensity at the cortex of MCF-7 cells after stimulation by laser ablation. (C) Levels of CTGF and NR4A3 transcripts in MCF-7 cells at the indicated times after addition of 1 µM ionomycin (at t = 0) as determined by quantitative RT-PCR (mean ± SD, n = 2, normalized against untreated control cells at each time point). (D) MCF-7 cells were pretreated with the indicated drugs (ROCK inhibitor Y27632, CaM-kinase inhibitors KN62 and KN93, calcineurin inhibitor cyclosporin A (CsA), actin inhibitors latrunculin A (LatA) and cytochalasin D (CytoD); concentrations are indicated in µM) for 10 min before addition of 1.5 µM ionomycin, and levels of CTGF transcripts were determined. Values are mean ± SD, n = 3. (E) Lifeact-GFP expressing MCF-7 cells were pretreated with the indicated inhibitors (CsA: 1 µM, Y27632: 10 µM, C3: 2.5 µg/ml) for at least 2 hr before the addition of 1 µM ionomycin to induce CaAR. Upper right: MCF-7 cells expressing Lifeact-GFP stimulated by the addition of 40% FBS and then monitored by fluorescence microscopy. Times in sec. Scale bars 10 µm.
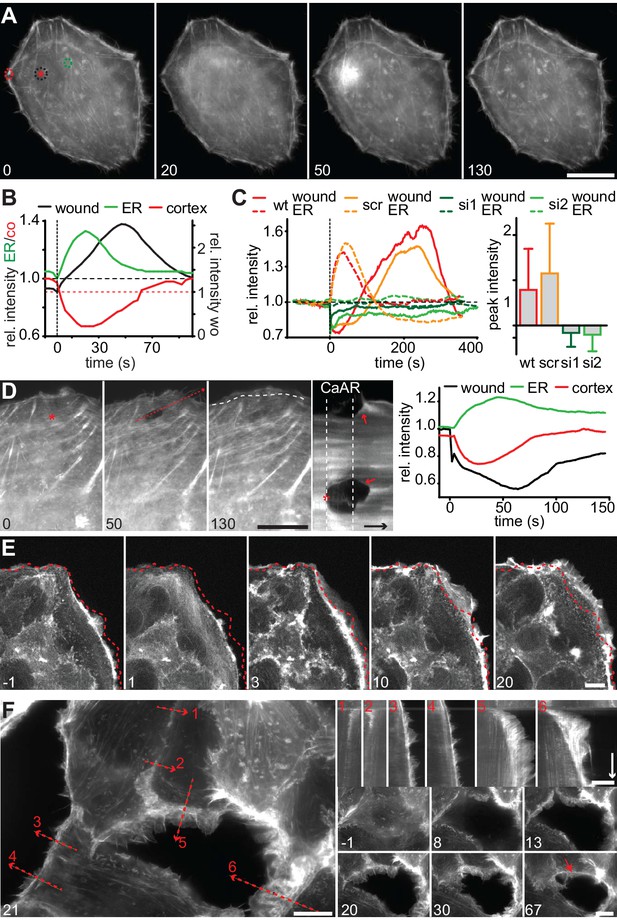
CaAR mediates acute cellular reorganization.
(A) A HeLa cell expressing Lifeact-GFP was damaged by laser ablation and Lifeact-GFP intensity was monitored in different regions. Asterisk: ablation site. Regions for intensity measurements in (B) are indicated in corresponding colors. (B) Plots of Lifeact-GFP signal intensity at ER, cortex and at the ablation site of the cell shown in (A). (C) Plots of Lifeact-GFP signal intensity in indicated regions of control and INF2-siRNA-treated cells. Quantification of peak intensities at the wounding site is shown as mean ± SD (n > 9). (D) A podocyte expressing Lifeact GFP was damaged by laser ablation and subsequently monitored by fluorescence microscopy. Asterisk indicates site of laser ablation. Red dotted line indicates the path of kymograph, a white dotted line indicates the outline of the cell before ablation, and arrows indicate instances of wound-repair and lamellipodia formation after CaAR completion. Lifeact-GFP intensity curves for ER, cortex and wound site are shown in the graph. Time arrow: 50 s. (E) Increase in cortical actin at the cell periphery upon activation of CaAR in MCF-7 cells with 50 µM ATP. The dotted lines indicate the position of the cell boundary before stimulation. (F) Changes in cortical actin dynamics after ablation of an MCF-7 cell within a monolayer. Kymographs are shown along the dotted lines for indicated positions (red numbers). Time arrow: 20 min. At later time-points actin congresses into ring structures around gaps (red arrow). Times in sec (A–D) and min (E, F). Scale bars: 10 µm.
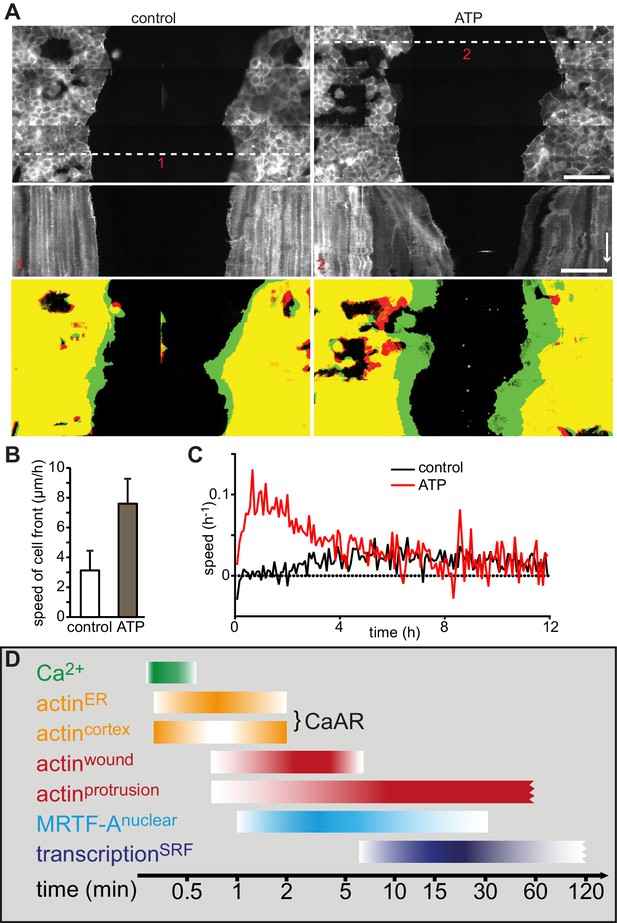
Wound healing following induction of CaAR and timeline.
(A) Representative examples of MCF-7 cells expressing Lifeact-mCherry seeded on either side of a PDMS spacer. After removal of the spacer cells were cultured for 24 hr and then treated with either control medium (left panels) or medium containing 50 µM ATP (right panels). Shown are the cell positions at the time of the medium exchange (offset resulting from stitching), the kymographs along the indicated lines (red numbers) and the overlay between t0 (red) and t5h (green). Time arrow: 4 hr. Scale bars: 100 µm. (B) Rate of cell front movement for control vs. ATP-treated cells. Student’s t-test p<0.05 for n = 5 experiments. (C) Graphs showing speed of closure in control cells vs. ATP-induced cells. (D) Timeline of CaAR and associated processes as discussed in the text.
Videos
MDCK cells expressing Lifeact-mCherry and labeled with Fluo4 exposed to shear flow (10 dyn/cm2).
Corresponds to Figure 1A. Scale bar: 10 µm.
A panel of indicated cell types expressing Lifeact-GFP exposed to 1 µM ionomycin.
Corresponds to Figure 1—figure supplement 1A. Scale bar: 10 µm.
MCF-7 cells expressing Lifeact-GFP exposed to 1 µM ionomycin.
Corresponds to Figure 1D. Scale bar: 10 µm.
HeLa cell expressing Lifeact-GFP labelled with Lysotracker Red and stimulated by laser ablation.
Corresponds to Figure 2E.
MCF-7 cell expressing Lifeact-GFP repeatedly stimulated by AFM (asterisk).
Corresponds to Figure 2—figure supplement 1D. Scale bar: 10 µm.
Propagation of CaAR in MCF-7 cells expressing Lifeact-GFP stimulated by laser ablation (asterisk).
Corresponds to Figure 2—figure supplement 1F. Scale bar: 10 µm.
MCF-7 cells expressing Lifeact-GFP, either untreated or pretreated with 400 nM LatA or 1 µM CytoD were exposed to 1 µM ionomycin at t = 0 s.
Corresponds to Figure 3A. Scale bar: 10 µm.
Cortical actin reorganization in an MCF-7 cell expressing Lifeact-GFP stimulated by laser ablation.
Corresponds to Figure 4—figure supplement 1A. Scale bar: 10 µm.
HeLa cell expressing Lifeact-GFP stimulated by laser ablation.
Corresponds to Figure 6A. Scale bar: 10 µm.
AB8 podocyte expressing Lifeact-GFP stimulated by laser ablation (asterisk).
Corresponds to Figure 6D. Scale bar: 10 µm.
Spreading of MCF-7 cells expressing Lifeact-GFP stimulated by ATP.
Corresponds to Figure 6E. Scale bar: 10 µm.
MCF-7 cells expressing Lifeact-GFP stimulated by laser ablation of a single cell in a monolayer.
Corresponds to Figure 6F. Scale bar: 10 µm.
MCF-7 cells expressing Lifeact-GFP migrating in to a gap in the absence (left) or presence (right) of 50 µM ATP.
Corresponds to Figure 7A.
Tables
Quantitative analysis of CaAR.
Cell type | MCF-7 | MCF-7 | MCF-7 | MCF-7 | HeLa | HeLa | |
|---|---|---|---|---|---|---|---|
20,74 | 14,09 | 8,20 | 11,78 | 15,59 | 5,34 | mean | |
increasetime constant (s) | 12,94 | 6,94 | 5,42 | 11,12 | 10,86 | 3,99 | stdev |
980 | 588 | 102 | 252 | 371 | 258 | n | |
**** | **** | **** | **** | **** | ANOVA | ||
−41,47 | −28,21 | −17,15 | −42,74 | −119,30 | −49,42 | mean | |
decreasetime constant (s) | 26,32 | 17,06 | 11,75 | 30,90 | 40,00 | 33,71 | stdev |
1010 | 586 | 102 | 279 | 136 | 193 | n | |
**** | **** | ns | **** | ns | ANOVA | ||
3,29 | 2,51 | 2,99 | 3,52 | 6,97 | 2,91 | mean | |
amplitudeintensity (a.u.) | 1,50 | 0,71 | 1,36 | 1,96 | 3,15 | 1,34 | stdev |
979 | 588 | 102 | 252 | 371 | 255 | n | |
**** | ns | ns | **** | *** | ANOVA | ||
217,30 | 137,00 | 72,65 | 162,70 | 391,40 | 115,20 | mean | |
plateautime (s) | 108,20 | 61,49 | 28,60 | 110,50 | 197,40 | 62,77 | stdev |
965 | 586 | 102 | 238 | 136 | 193 | n | |
**** | **** | **** | **** | **** | ANOVA |
Model Input parameters.
Parameter | Symbol | Value | Reference |
|---|---|---|---|
maximum polymerization speed | 15.6 μm s-1 | ||
depolymerization speed | 0.1 μm s-1 | ||
capping rate | 3 s-1 | ||
severing rate | 0.005 μm-1s-1 | ||
maximum cortical nucleation rate | 100 s-1 | estimated | |
maximum ER nucleation rate | [500–1100] s-1 | estimated | |
length scale | 2000 μm | assumption | |
total available actin | 4.0 | estimated |
Additional files
-
Source code 1
Matlab script for automated nuclear rim detection.
- https://doi.org/10.7554/eLife.19850.032
-
Source code 2
Matlab script for manual nuclear rim detection.
- https://doi.org/10.7554/eLife.19850.033
-
Source code 3
Matlab script for analysis of intensity traces at nuclear rims.
- https://doi.org/10.7554/eLife.19850.034





