The fail-safe mechanism of post-transcriptional silencing of unspliced HAC1 mRNA
Figures
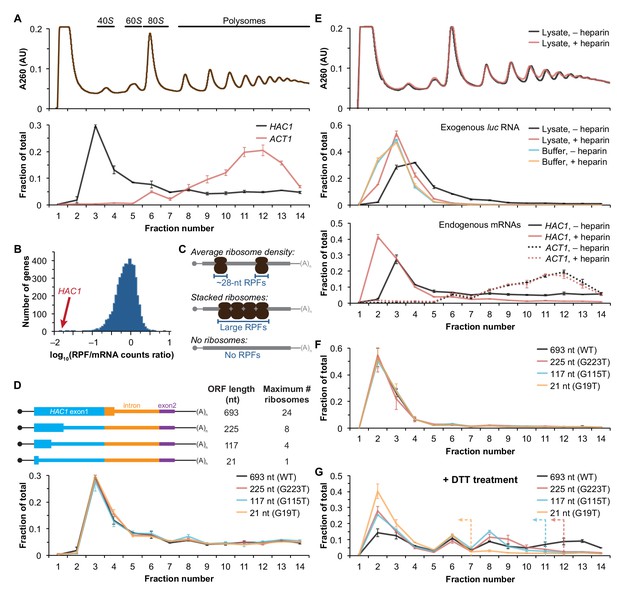
Ribosome density on unspliced HAC1 mRNA.
(A) Polysome analysis of HAC1 and ACT1 mRNAs. Extracts prepared from exponentially growing yeast cells were fractionated on 10–50% sucrose gradients, with absorbance at 260 nm monitored (top). The relative distributions of HAC1 and ACT1 mRNAs across fractions were determined by qRT-PCR (bottom). Shown are the mean ± SEM with n = 2 (i.e., the range), expressed as a fraction of the total mRNA detected. (B) Histogram of ribosome densities measured by ribosome profiling and RNA-seq. The ratio of the number of ribosome-protected fragments (RPFs) to the number of RNA-seq reads (mRNA counts) was calculated for each of 4838 expressed yeast genes (data from Weinberg et al., 2016). Shown is the distribution of log-transformed ratios in bins of 0.05, with the position of HAC1 indicated. (C) Possible scenarios to explain a lack of RPFs. While an mRNA with average ribosome density will generate many ~28 nucleotide (nt) RPFs (top), the close packing of stacked ribosomes could inhibit the RNase digestion between ribosomes required to generate ~28 nt RPFs (middle). Alternatively, an mRNA that does not contain translating ribosomes would not generate RPFs (bottom). (D) Polysome analysis of HAC1 mRNA variants with shortened ORFs. G-to-T point mutations were introduced into the first exon of HAC1 to generate premature stop codons, with the resulting ORFs shown as thick colored boxes (constitutive 5′- and 3′-UTRs located within exons 1 and 2, respectively, are shown as thin black lines; other untranslated regions are depicted as thin colored boxes; and the coding regions of exons 1 (teal) and 2 (purple) are labeled as 'HAC1 exon1' and ‘exon2’, respectively). The maximum number of ribosomes that could be accommodated was calculated based on each ribosome occupying 28 nt. Polysome analysis was performed as in (A), with data for wild-type HAC1 from (A) duplicated for comparison. (E) Effects of heparin on polysome analysis. Purified uncapped luciferase (luc) RNA was added to either lysate or lysis buffer in the absence (–) or presence (+) of 0.2 mg/ml heparin. Polysome analysis was performed as in (A) with absorbance at 260 nm monitored (top), and the relative distributions of exogenous luc RNA (middle) and endogenous HAC1 and ACT1 mRNAs (bottom; in lysate only) were determined. (F) Refined polysome analysis of HAC1 mRNAs. Extracts were prepared in heparin-containing lysis buffer from strains shown in (D). Polysome analysis was performed as in (A). (G) Polysome analysis of HAC1 mRNAs during the UPR. Strains shown in (D) were grown to mid-log phase and treated with 8 mM DTT for 20 min before harvesting. Extracts were prepared in heparin-containing lysis buffer, and polysome analysis was performed as in (A). Dotted lines indicate the fractions after which the corresponding color-coded mutant mRNAs would not be expected to sediment based on ORF length.
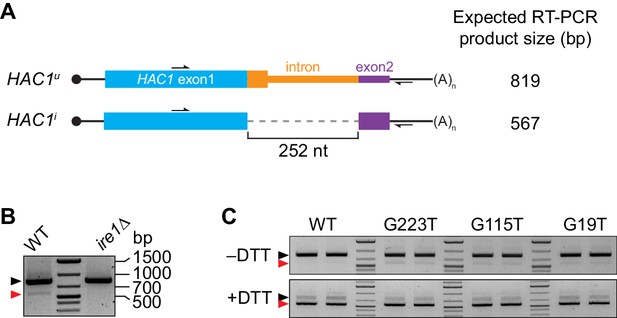
Splicing of HAC1 mRNA.
(A) Design of RT-PCR assay for HAC1 splicing. To differentiate between the HAC1u and HAC1i mRNAs, reverse-transcription products are PCR amplified using a pair of primers that flank the intron. The resulting PCR products are separated by agarose gel electrophoresis, with a size difference of 252 base pairs (bp) corresponding to the size of the intron. (B) Splicing status of HAC1 mRNA. Total RNA was extracted from exponentially growing yeast cells of the indicated genotypes. RT-PCR across the intron was used to differentiate between unspliced (black arrow) and spliced (red arrow) HAC1 mRNA. The small percentage of spliced HAC1 mRNA observed in wild-type cells is IRE1 dependent, as expected. (C) Splicing of HAC1 mRNA variants upon DTT treatment. Strains expressing the indicated HAC1 variants were grown to mid-log phase and either left untreated (–DTT) or treated with 8 mM DTT for 20 min (+DTT) before harvesting. Total RNA was extracted and used for RT-PCR analysis as in (A). The same cultures (two of each strain) were used for polysome analysis in Figure 1F and G.
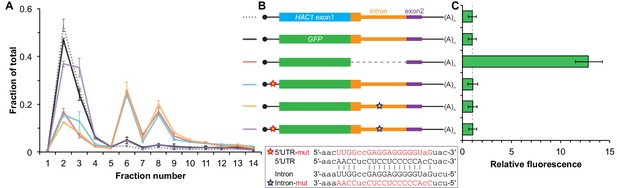
Contribution of long-range base pairing to intron-dependent silencing.
(A) Polysome analysis of reporter mRNAs. Extracts were prepared in heparin-containing lysis buffer from strains expressing the GFP reporter mRNAs depicted in (B). Polysome analysis was performed as in Figure 1A, with data for wild-type HAC1 from Figure 1F duplicated for comparison. (B) Design of reporter mRNAs. Constructs are depicted as in Figure 1D, with the dotted line indicating a deleted region. Colored stars indicate mutations to the base-pairing region, with specific nucleotide changes shown below in red. (C) Flow cytometry analysis of reporter strains. Strains expressing the GFP reporter mRNAs depicted in (B) were grown to mid-log phase and analyzed by flow cytometry. Plotted is the median GFP intensity (normalized to cell size) of the cell population relative to background fluorescence in the wild-type (no GFP) strain with error bars indicating quartiles of the cell population, all averaged across replicates (n = 2–7).
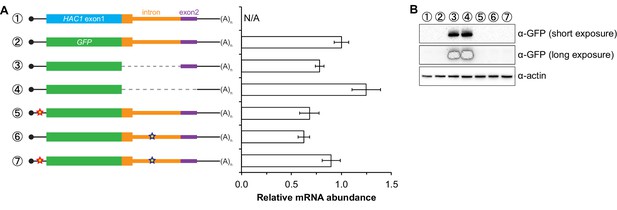
Characterization of GFP reporter strains.
(A) RNA abundance measurements for GFP reporter mRNAs. Total RNA was extracted from strains expressing the indicated mRNAs, using the same cultures as for flow cytometry analyses in Figure 2C. qRT-PCR was used to measure the abundances of GFP reporter mRNAs relative to ACT1 mRNA, with all data normalized to the abundance of the ‘wild-type’ GFP reporter mRNA (construct 2). Shown are the mean ± SD (n = 2–7). (B) Immunoblotting analysis of reporter strains. Proteins were extracted from mid-log-phase cultures of strains expressing the indicated reporter mRNAs depicted in (A). Immunoblots for the GFP reporter and actin loading control are shown. No signal for GFP is observed in the base-pairing mutants (constructs 5 and 6) even after overexposing the blot, consistent with the corresponding flow cytometry results (Figure 2C).
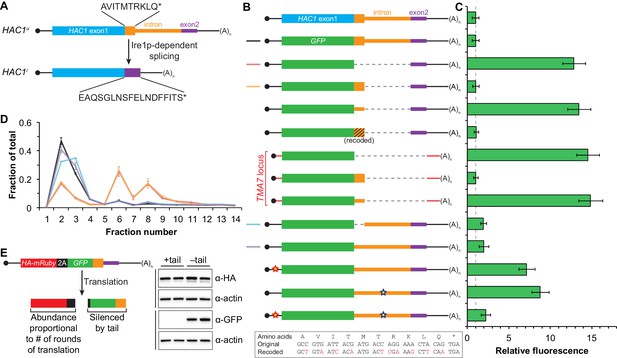
Post-translational silencing mediated by the intron-encoded C-terminal tail.
(A) Schematic of HAC1 mRNA splicing. The proteins encoded by HAC1u and HAC1i mRNAs differ in their C-terminal tails, with the amino acid sequences indicated. (B) Design of reporter mRNAs. Black shading indicates recoding, with the original and recoded sequences depicted below (mutations in red). Untranslated regions colored red correspond to those of the TMA7 mRNA, with the reporter gene integrated at the TMA7 rather than HAC1 locus. Otherwise constructs are depicted as in Figure 2B. (C) Flow cytometry analysis of reporter strains. Strains expressing the GFP reporter mRNAs depicted in (B) were analyzed as in Figure 2C, with data for the first three strains duplicated from Figure 2B for comparison. (D) Polysome analysis of reporter mRNAs. Extracts were prepared in heparin-containing lysis buffer from strains expressing the GFP reporter mRNAs indicated in (B). Polysome analysis was performed as in Figure 1A, with data for the wild-type and intronless GFP reporters from Figure 2A duplicated for comparison. (E) Differentiating between co-translational and post-translational silencing mechanisms. Left: Schematic of reporter construct that generates two separate polypeptides from each round of translation. Right: Extracts were prepared from strains expressing reporter mRNAs that either encoded the 10-amino-acid C-terminal tail of Hac1up (+tail) or contained a stop codon just before the tail (–tail). Immunoblotting was used to detect HA-tagged mRuby (top) and GFP (bottom), with actin as a loading control. Two biological replicates are shown for each genotype.
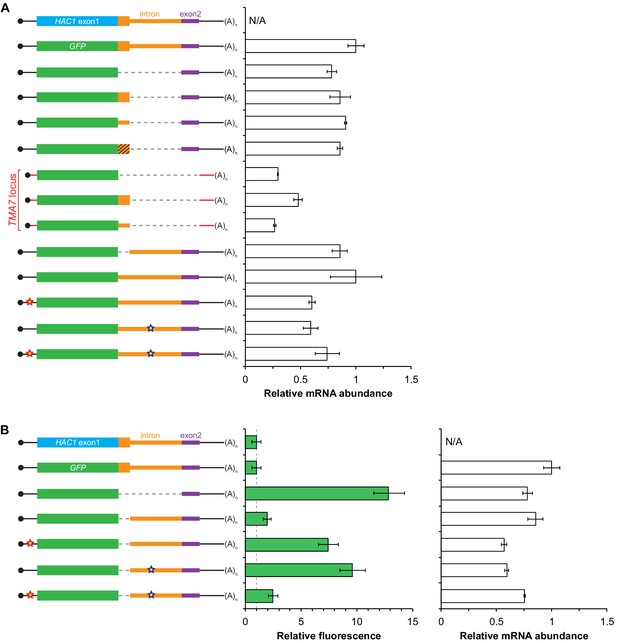
Additional analyses of GFP reporter constructs.
(A) RNA abundance measurements for GFP reporter mRNAs, analyzed as in Figure 2—figure supplement 1A using the same cultures as for flow cytometry analyses in Figure 3C. (B) Flow cytometry and RNA analyses of additional reporter strains. Strains expressing the indicated GFP reporter mRNAs (left) were analyzed by flow cytometry as in Figure 2C (middle) or qRT-PCR as in Figure 2—figure supplement 1A (right), with data for the first four strains duplicated from Figure 3C and Figure 3—figure supplement 1A for comparison.
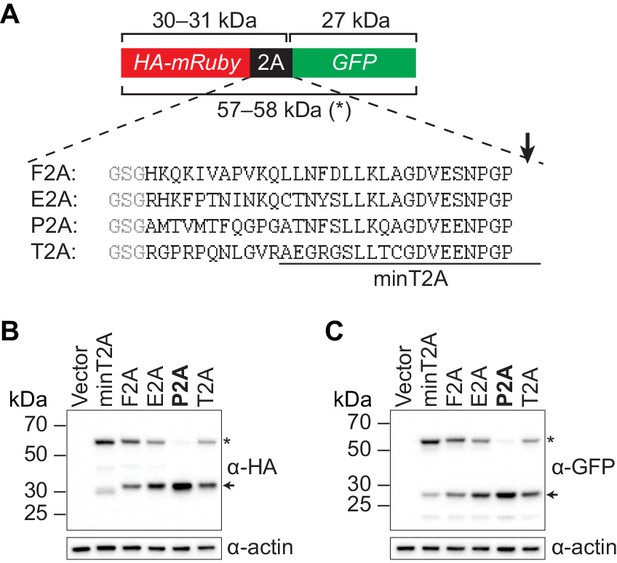
Identifying a 2A peptide sequence that is active in S. cerevisiae.
(A) Design of the 2A reporter construct. In the absence of 2A activity, a 57–58 kDa polypeptide (*) is produced that contains both an HA tag and GFP. When the 2A sequence is active, translation of the reporter generates a 30–31 kDa HA-tagged protein and a 27 kDa GFP. Shown are the 30-amino-acid sequences of the 2A peptides that were tested, with each preceded by a GlySerGly linker (gray) and ‘cleaved’ at the site marked with an arrow. The region corresponding to a minimal 20-amino-acid T2A peptide (‘minT2A’) that has been used previously in yeast (Beekwilder et al., 2014) is indicated. Minimal versions of the F2A peptide have also been used in yeast (Doronina et al., 2008; de Felipe et al., 2003; Sharma et al., 2012; Voordeckers et al., 2012) but with ‘cleavage’ efficiencies that were low or not directly measured. (B–C) Analysis of 2A activity by immunoblotting. BY4741 was transformed with a pRS413-derived centromeric plasmid containing the indicated reporter construct under control of the GPD promoter. Proteins were extracted from mid-log-phase cultures and analyzed for HA- (B) and GFP- (C) tagged proteins by immunoblotting. Only the P2A peptide causes efficient separation of the upstream and downstream proteins (indicated by arrows). The nucleotide sequences encoding the 30-amino-acid peptides are:
F2A: CACAAACAAAAGATTGTTGCGCCTGTGAAACAGCTTTTGAACTTTGACCTGCTCAAGTTGGCAGGAGACGTCGAGTCCAACCCTGGGCCT
E2A: AGACATAAATTTCCCACTAACATCAACAAACAGTGTACTAATTACTCTCTCCTCAAATTGGCTGGAGATGTTGAGAGCAACCCTGGC CCC
P2A: GCTATGACTGTGATGACATTCCAGGGACCAGGTGCAACAAACTTCTCCCTCTTGAAACAAGCAGGAGATGTTGAGGAAAATCCCGGGCCT
T2A: CGGGGGCCTCGCCCCCAAAACCTTGGGGTAAGGGCCGAGGGCAGGGGAAGTCTTCTAACATGCGGGGACGTGGAGGAAAATCCCGGCCCC
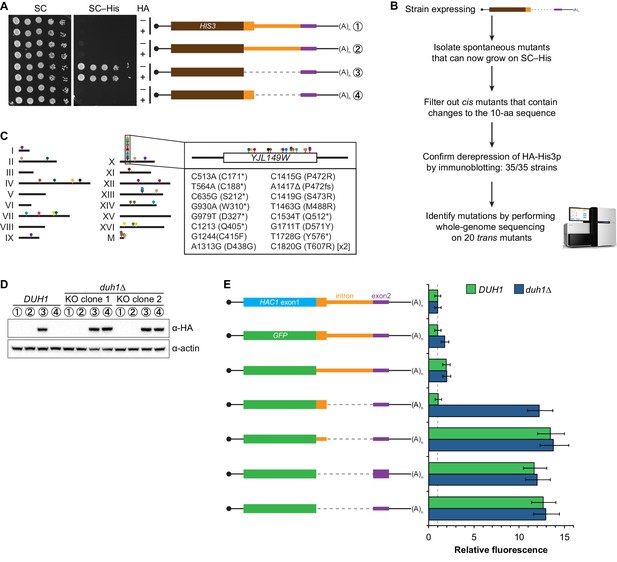
Identification of DUH1 through a genetic selection.
(A) Evaluating a genetic reporter for intron-dependent silencing. Strains expressing the indicated HAC1-based HIS3 reporter mRNAs (depicted as in Figure 2B) either without (–) or with (+) an N-terminal HA tag were grown to saturation, and 10-fold dilution series were plated on either SC or SC–His media. (B) Flowchart of genetic selection for dds mutants. After selecting for spontaneous mutants that could grow on medium lacking histidine, restreaked clones were filtered out for cis mutants, verified to be expressing HA-His3p, and a subset analyzed by whole-genome sequencing. (C) Chromosome map of mutations identified by whole-genome sequencing. Each color corresponds to a different dds mutant strain. Shown on the right is the DUH1 locus (YJL149W), with locations of nucleotide changes (with respect to DUH1 start codon) and corresponding amino changes listed below. (D) Effect of DUH1 disruption on expression of the genetic reporter. Strains expressing the indicated reporter mRNAs depicted in (A) in either a DUH1 or duh1∆ (two independent clones) background were grown to mid-log phase. Extracts were prepared and immunoblotted for HA-His3p and actin loading control. (E) Flow cytometry analysis of reporter strains. Strains expressing the indicated GFP reporter mRNAs in either a DUH1 or duh1∆ background were analyzed as in Figure 2C. Data is plotted as in Figure 2C (n = 2 for all strains).
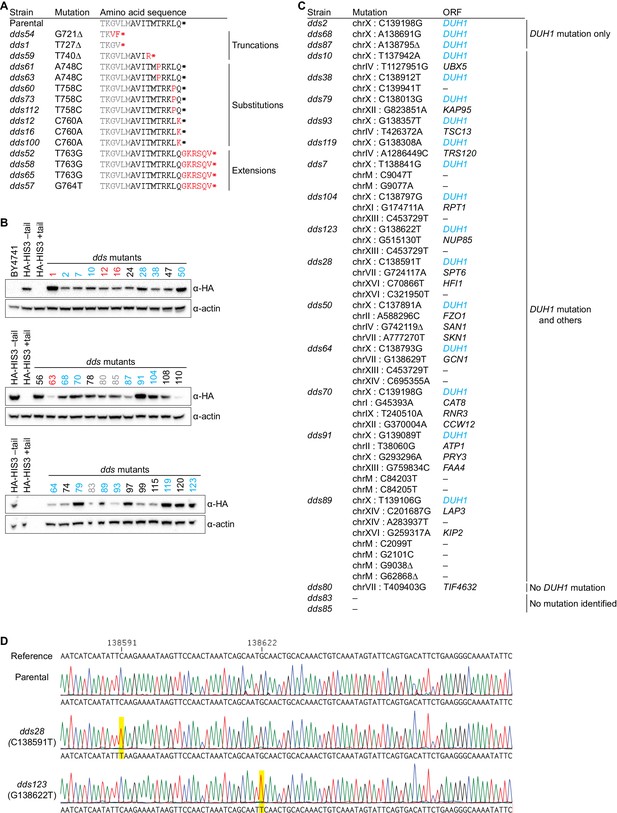
Results and validation of the genetic selection.
(A) Table of cis-acting mutations identified by Sanger sequencing. In the parental strain, the Hisp3-coding sequence is shown in gray while the 10-amino-acid degron is shown in black. Amino-acid changes caused by nucleotide mutations are shown in red. (B) Analysis of HA-His3p abundance in dds mutant strains. Protein extraction and immunoblotting were performed as in Figure 4D, using untagged BY4741 as a negative specificity control for the HA antibody. Strains indicated in red contain mutations in the HA-His3p +tail reporter gene. Strains that were subjected to whole-genome sequencing are indicated in blue and gray, with blue indicating those that contain a mutation in DUH1 and gray indicating those that do not. (C) Table of trans-acting mutations identified by whole-genome sequencing. Strains containing DUH1 mutations are listed in order of increasing total number of mutations. (D) Representative example of Sanger sequencing confirmation of DUH1 mutations. Shown are sequencing traces across a region of DUH1 for the parental selection strain and two dds mutants. Each dds mutant contains a nucleotide substitution (highlighted in yellow), compared to both the reference sequence and parental selection strain, corresponding to the mutation identified by whole-genome sequencing.
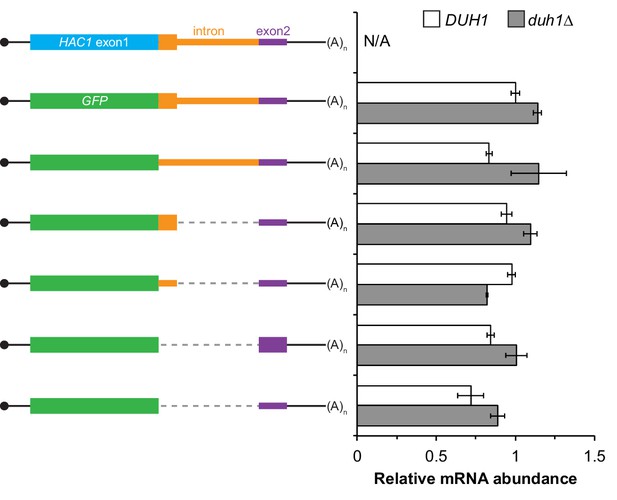
Additional analysis of GFP reporter constructs.
RNA abundance measurements for GFP reporter mRNAs in the indicated strain backgrounds, analyzed as in Figure 2—figure supplement 1A using the same cultures as for flow cytometry analyses in Figure 4E.
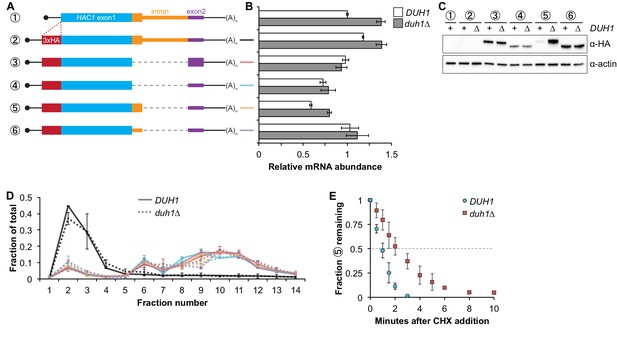
Effects of DUH1 on expression and stability of Hac1p.
(A) Design of 3xHA-tagged HAC1 mRNA variants. Constructs are depicted as in Figure 2B, with the location of the N-terminal 3xHA tag indicated. (B) RNA abundance measurements for HAC1 mRNA variants. Total RNA was extracted from strains expressing the indicated mRNAs in either a DUH1 or duh1∆ background. qRT-PCR was used to measure the abundances of HAC1 variants relative to ACT1 mRNA, with all data normalized to the abundance of construct 1 in strain BY4741. Shown are the mean ± SD (n = 2). (C) Effect of DUH1 disruption on protein abundances. Strains expressing the indicated mRNAs depicted in (A) in either a DUH1 or duh1∆ background were grown to mid-log phase. Extracts were prepared and immunoblotted for 3xHA-Hac1p and actin loading control. (D) Polysome analysis of 3xHA-tagged HAC1 mRNA variants. Extracts were prepared in heparin-containing lysis buffer from strains expressing the mRNAs indicated in (A) in either a DUH1 or duh1∆ background. Polysome analysis was performed as in Figure 1A. (E) Analysis of protein degradation kinetics. Strains expressing construct 5 (depicted in A) in either a DUH1 or duh1∆ background were grown to mid-log phase before being treated with cycloheximide (CHX) to halt translation. At the indicated time points, aliquots of cells were quenched in dry-ice-cold methanol and harvested by centrifugation. Protein extraction and immunoblotting were performed as in (C), except that a high-sensitivity antibody was used to detect 3xHA-Hac1up. Shown are the mean ± SD (n = 3), expressed as a fraction of protein detected at t = 0.

Relationship between base pairing– and degron-dependent repression.
(A) Design of 3xHA-tagged HAC1 mRNA variants. Constructs are depicted as in Figure 2B. Colored stars indicate mutations to the base-pairing region, with specific nucleotide changes shown in Figure 2B (red and blue stars) or below (green and black stars) in red. (B) RNA abundance measurements for HAC1 mRNA variants in the indicated strain backgrounds, analyzed as in Figure 5B. (C–D) Effect of DUH1 disruption on protein abundances. ire1∆ strains expressing the indicated mRNAs depicted in (A) in either a DUH1 or duh1∆ background were analyzed as in Figure 5C, except that a high-sensitivity antibody was used to detect 3xHA-Hac1up.
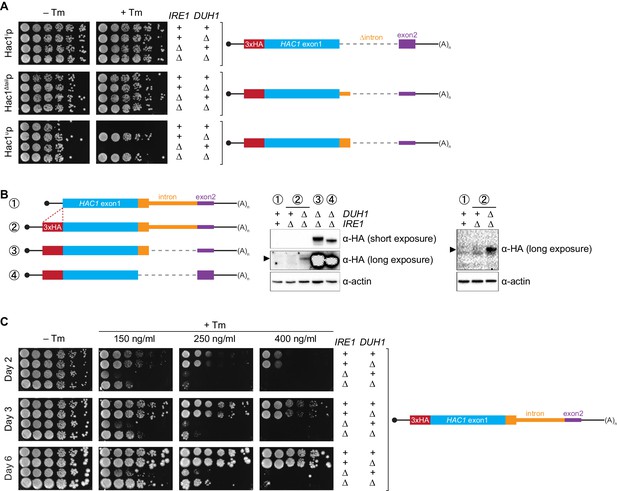
Requirement for DUH1 to suppress Ire1p-independent activation of the UPR.
(A) Analysis of Hac1p activity in the UPR. Strains expressing the indicated HAC1 mRNA variants, with (+) or without (∆) IRE1 and/or DUH1 present, were grown to saturation. 10-fold dilution series were plated on YPD without (–Tm) or with (+Tm) 400 ng/ml tunicamycin to induce ER stress. (B) Impact of DUH1 on detection of Hac1up. Strains expressing the indicated mRNAs, with (+) or without (∆) IRE1 and/or DUH1 present, were analyzed as in Figure 6C. Black arrow indicates the position of Hac1up, which migrates more slowly than Hac1ip. Construct 1, which lacks a 3xHA tag, was used as a negative control for anti-HA immunoblotting. (C) Effect of Duh1p-dependent degradation on the UPR. Strains expressing wild-type HAC1 with an N-terminal 3xHA tag, with (+) or without (∆) IRE1 and/or DUH1 present, were grown to saturation. 10-fold dilution series were plated on YPD without tunimacyin (–Tm) or containing the indicated concentration of tunicamycin (+Tm). Plates were imaged at days 2 (top), 3 (middle), and 6 (bottom).
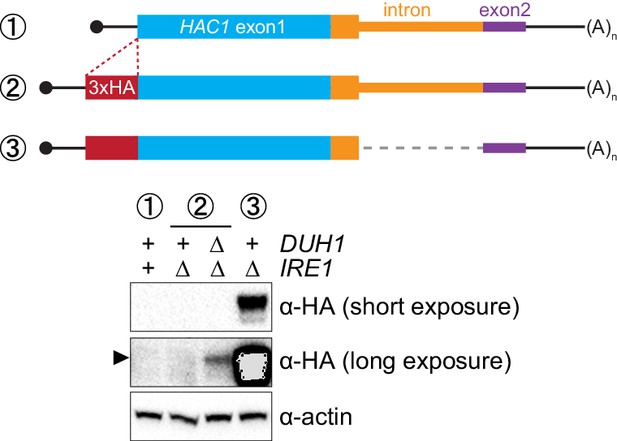
Ire1p-independent accumulation of Hac1p.
Strains expressing the indicated mRNAs, with (+) or without (∆) IRE1 and/or DUH1 present, were analyzed and presented as in Figure 7B.
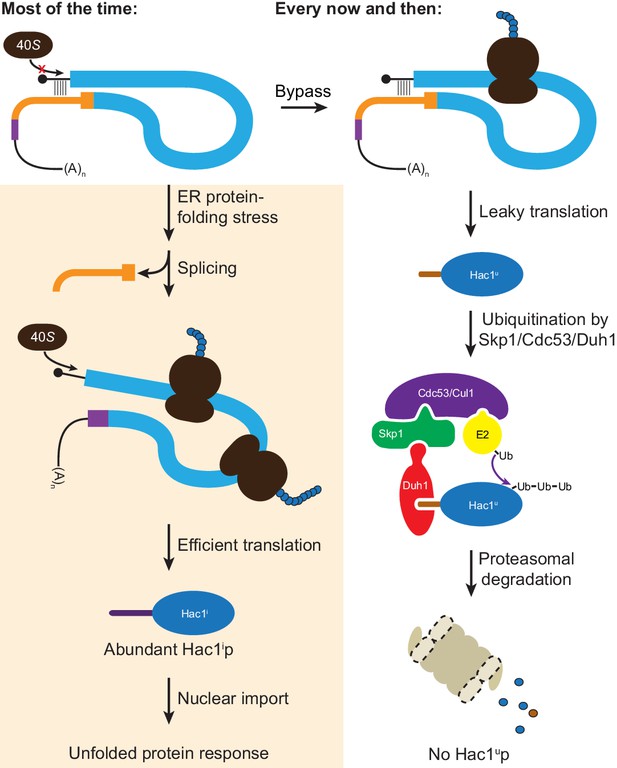
Fail-safe post-transcriptional silencing of unspliced HAC1 mRNA.
See the main text for a description.
Additional files
-
Supplementary file 1
Yeast strains used in this study.
See key below.
- https://doi.org/10.7554/eLife.20069.018





