Klp10A, a stem cell centrosome-enriched kinesin, balances asymmetries in Drosophila male germline stem cell division
Figures
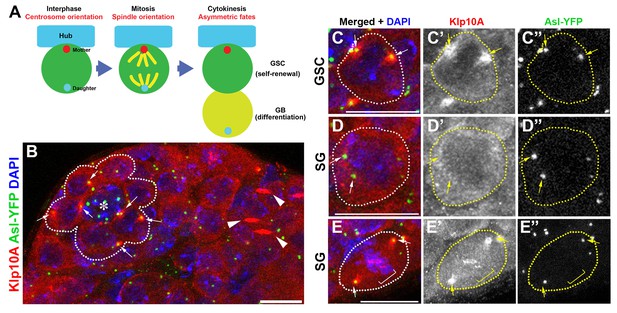
Klp10A localizes to the GSC centrosomes.
(A) Centrosome behavior in male GSCs. (B) An apical tip of the testis stained for Klp10A (red), Asl-YFP (green, centrosome) and DAPI (blue). Arrows indicate GSC centrosomes. GSCs are indicated by broken lines. Arrowheads indicate Klp10A localization to the central spindle microtubule bundle. Hub (*). Bar: 10 µm. (C, D) magnified images of an interphase GSC (C) and an interphase spermatogonium (D) chosen from panel B, demonstrating that Klp10A localization to the interphase centrosome is specific to GSCs. E) Mitotic spermatogonium showing Klp10A localization to the spindle poles (arrows) and kinetochores (bracket). Bar: 5 µm. It should be noted that MT-nanotubes are sensitive to regular fixation conditions, and its localization on MT-nanotubes is not obvious in the images shown in panel B.
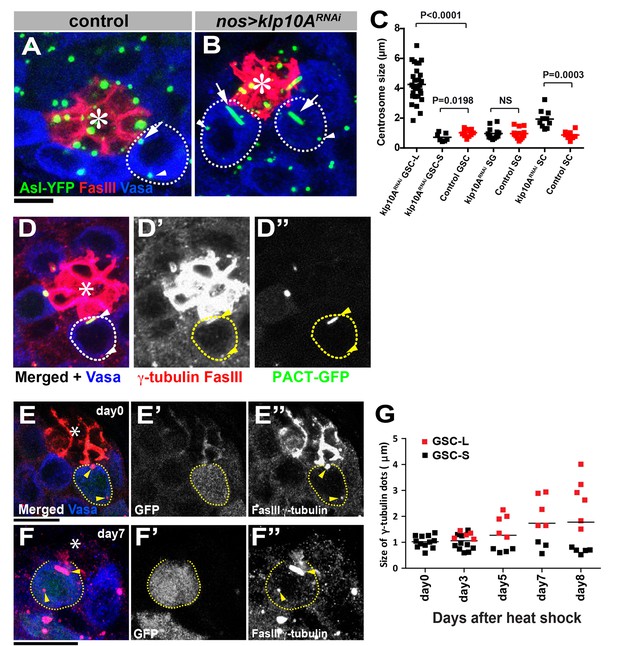
klp10A is required for maintenance of mother centrosome size in GSCs.
Wild-type (A) and klp10ARNAi (B) GSCs stained for Asl-YFP (green, centrosome), Fas III (red, hub), Vasa (blue). GSCs are indicated by dotted circles. Arrows indicate proximal (mother) centrosomes; arrowheads indicate distal (daughter) centrosomes. Hub (*). Bar: 5 µm. (C) Quantification of centrosome size in control (N = 13) and klp10ARNAi (N = 30 GSC-L, N = 15 GSC-S) GSCs, control (N = 14) and klp10ARNAi (N = 15) SGs, and control (N = 9) and klp10ARNAi (N = 11) spermatocytes (SC). (D) PACT-GFP (NGT40>PACT-GFP) labels the elongated, proximal centrosome. γ-tubulin and Fas III (red), Vasa (blue), GFP-PACT (green). Hub (*). GSC is indicated by broken line, and the centrosomes are indicated by arrowheads. (E, F) Examples of klp10ARNAi GSC clones at 0 days (E) and seven days (F) after clone induction (hs-FLP, nos>stop>gal4, UAS-GFP, UAS-klp10ARNAi). GFP (green, klp10ARNAi GSC clones), Fas III, γ-tubulin (red). Bar: 10 µm. (G) Quantification of centrosome length in klp10ARNAi GSC clones on indicated days after clone induction. Day0 (N = 13), Day3 (N = 16), Day5 (N = 10), Day7 (N = 9) and Day8 (N = 12) were scored. Statistical analysis and graphing were performed using GraphPad prism six software. The p value (two-tailed Student’s t-test) is provided for comparison between indicated columns in C. Bars in G indicate means. Note that GSC-L and –S indicate longer and shorter centrosomes, respectively, from each GSC in C and G. Scoring of GSC centrosome size in these panels come only from GSCs whose centrosomes shows clear size difference (overall average size including GSCs without visibly elongated centrosomes is given in the text).
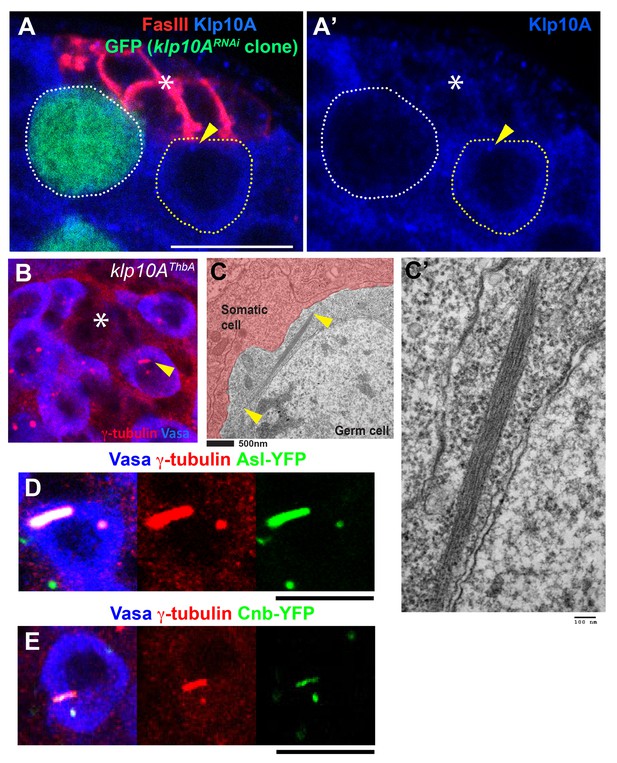
Loss of Klp10A leads to mother centrosome elongation.
(A) Validation of klp10ARNAi. klp10ARNAi GSC is marked by GFP (green) (hs-FLP, nos>stop>gal4, UAS-GFP, UAS-klp10ARNAi), and the testis was stained for anti-Klp10A (blue), and FasIII (red). Hub (*). klp10ARNAi GSC clone is indicated by white dotted line, and control GSC is indicated by yellow dotted line with arrowhead pointing the centrosome. (B) Centrosome elongation observed in testis of L3 larvae of klp10AThbA mutant (arrowhead). γ-tubulin (red), Vasa (blue). (C) Transmission electron microscopic image showing elongated centrosome in germ cell upon klp10ARNAi. Arrowheads indicate two ends of the centrosome visible in the EM section. Bar: 500 nm in C and 100 nm in C’. (D, E) Centrosomes in klp10ARNAi GSCs marked by Asl-YFP (D) or Cnb-YFP (E), stained for Vasa and γ-tubulin. Bar: 10 µm.
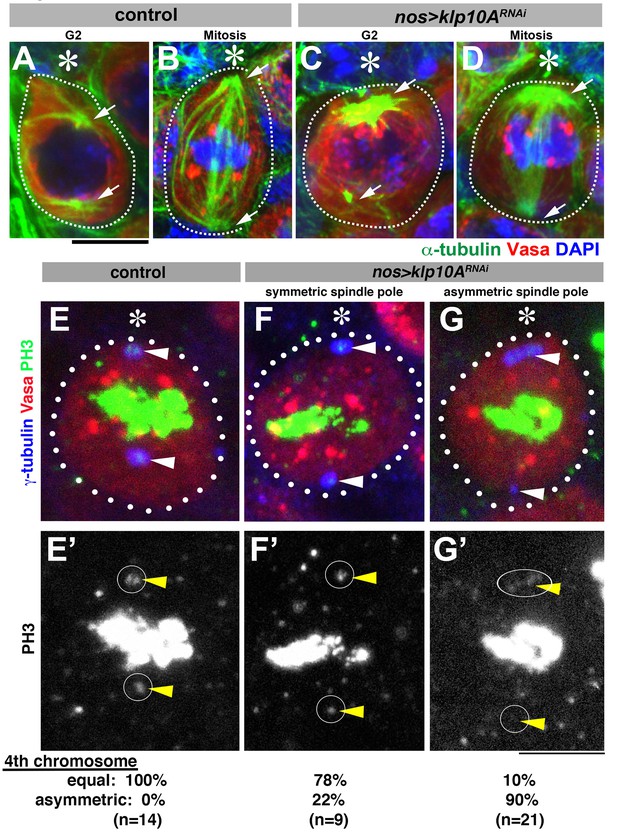
Depletion of klp10A results in unequal microtubule-organizing activity of the mother and daughter centrosomes.
(A–D) GSCs stained for α-tubulin (green), Vasa (red) and DAPI in control G2 phase (A), control mitosis (B), klp10ARNAi G2 phase (C) and klp10ARNAi mitosis (D). GSCs are indicated by dotted lines. Centrosomes/spindle poles are indicated by arrowheads. Hub (*). Bar: 5 µm. (E–G) Mitotic GSCs stained for Vasa (red), γ-tubulin (blue), and phosphor-histone H3 (PH3, green) in control GSC (E, E'), klp10ARNAi GSC with centrosomes of equal size (F, F'), and klp10ARNAi GSC with elongated mother centrosome (G, G'). The position of spindle pole, where chromosome 4 should be observed during metaphase, is indicated by arrowheads and circles.
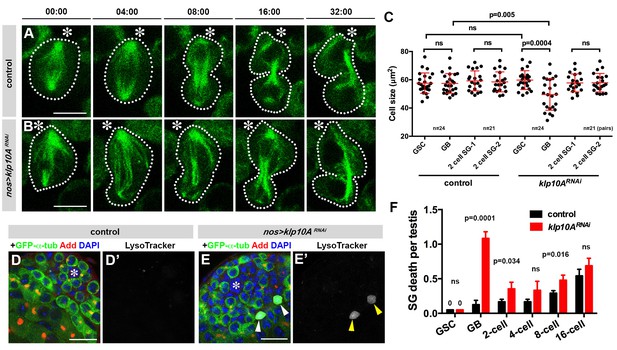
Depletion of klp10A generates GSC daughters with unequal cell sizes.
(A, B) Frames from time-lapse live observation of control (A) and klp10ARNAi (B) GSCs expressing GFP-α-tubulin. GSCs are outlined by dotted lines. Time is indicated in minutes. Movies were started during late metaphase, ~4 min prior to spindle elongation. Hub (*).Bar: 5 µm. (C) Quantification of cell size in GSCs and GBs or two SGs at the completion of GB mitosis to become 2-cell SGs in control vs. klp10ARNAi testes. N = 24 mitotic GSCs and N = 21 mitotic GBs were scored in both control and klp10ARNAi testes. (D, E) testis apical tip stained for GFP-α-tubulin (green), Adducin-like (red), DAPI (blue) and Lysotracker (white). Hub (*). Dying GB is indicated by arrowheads in E). Bar: 20 µm. F) Quantification of germ cell death in control vs. klp10ARNAi testes. N = 96 testes were scored in both control and klp10ARNAi for quantification. p value was obtained with Student’s t-test (two-tailed). Data are shown as mean ± s.d.
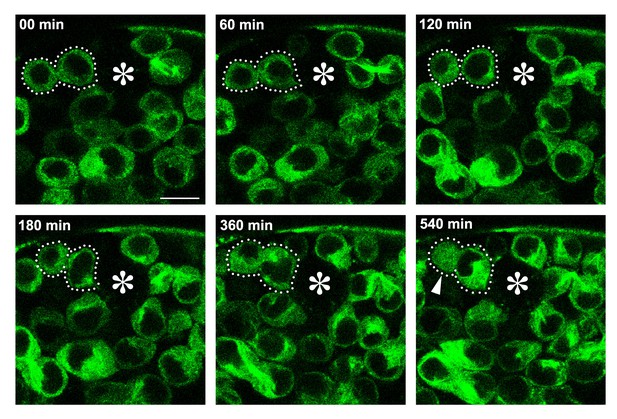
An example of GB death following klp10ARNAi GSC division with unequal daughter cell size.
klp10ARNAi GSC expressing GFP-α-tubulin (nos>UAS-GFP-α-tubulin, UAS-klp10ARNAi) that completed its mitosis (dotted lines) was followed by time-lapse live observation. At 540 min, GB was observed to have lost its nuclear integrity, in which normally cytoplasmic GFP-α-tubulin was observed throughout the cell (arrowhead). Bar: 10 µm.
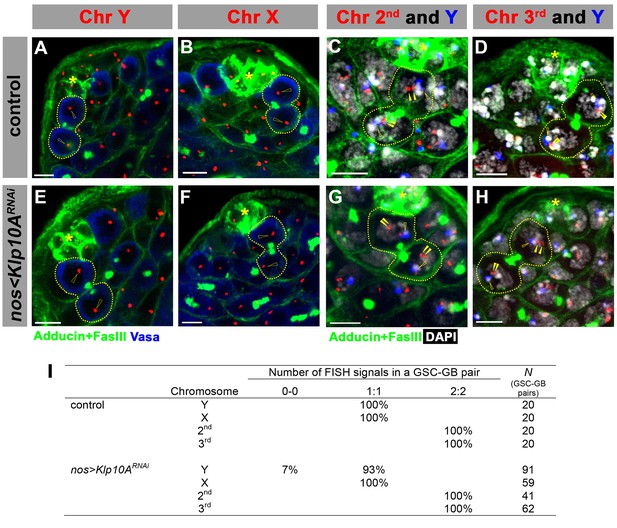
Segregation of major chromosomes is not affected in klp10ARNAi testes.
(A–H) Examples of chromosome segregation patterns in post mitotic GSC-GB pairs in control (A–D) and in klp10ARNAi (E–H) testes. Asterisks indicate the hub, and dotted lines indicate GSC-GB pairs. See Materials and methods for sequence information for FISH probes. Open arrowheads indicate sex chromosomes, and solid arrowheads indicate autosomes. Bars: 5 m. (I) Summary of sister chromatid segregation patterns. For sex chromosomes, 1:1 is the normal segregation pattern, and for the autosomes, 2:2 is the normal segregation pattern. Sister chromatid segregation was mostly normal in klp10ARNAi GSCs, expect for infrequent loss of Y chromosome.
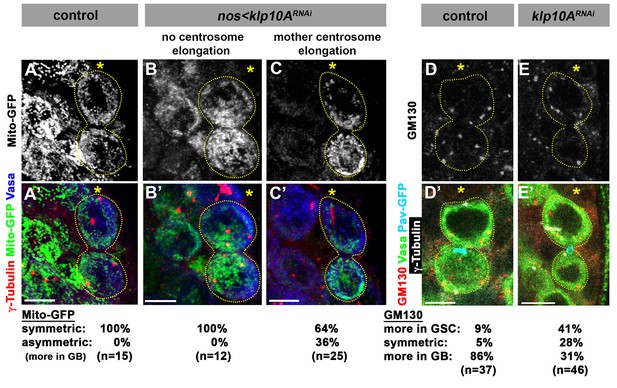
Segregation of mitochondria and Golgi is affected in klp10ARNAi testes.
(A–C) Mito-GFP distribution in control (A) and klp10ARNAi (B, C) GSC-GB pairs. Red: γ-tubulin. Green: mito-GFP. Blue: Vasa. (D, E) GM130 (Golgi) distribution in control (D) and klp10ARNAi (E) GSC-GB pairs. Asterisks indicate the hub, and dotted lines indicate GSC-GB pairs. Bars: 5 µm.





