Pharmacological targeting of the transcription factor SOX18 delays breast cancer in mice
Figures
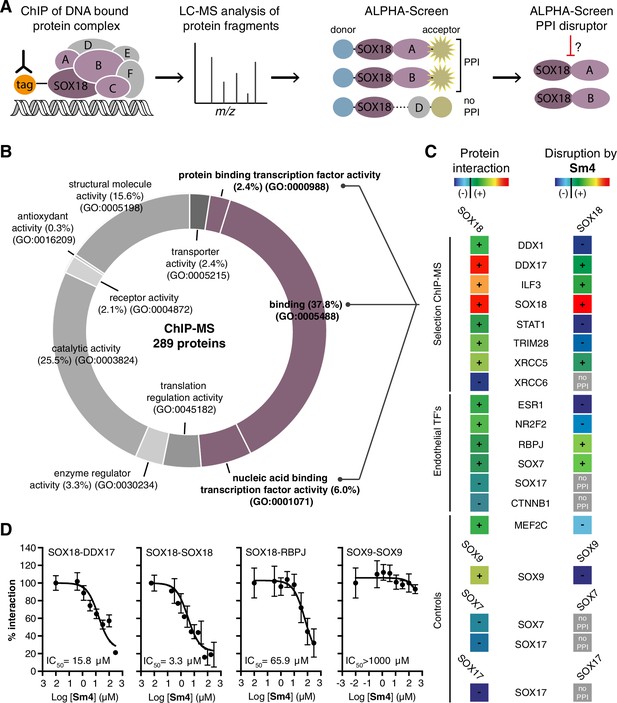
Mapping of SOX18 interactome and disruption of interactions by Sm4.
(A) Schematic of the experimental strategy to deconvolute SOX18-dependent protein-protein interactions (PPIs) combining Chromatin immunoprecipitation-mass spectrometry (ChIP-MS) and Amplified Luminescent Proximity Homogeneous Assay (ALPHA-Screen) methods. (B) GO-term analysis for molecular function on the 289 proteins identified by SOX18-cMyc ChIP-MS in human umbilical vein endothelial cells (HUVECs). Non-specific interactors found in Myc-tag only transfected cells were subtracted. Proteins with nucleic acid binding or protein binding capacity (purple) were considered for consecutive direct interaction studies to enhance likeness of identifying direct interactors. (C) Left column: heatmap representation of SOX18 pairwise PPIs as tested by ALPHA-Screen, on a selection of ChIP-MS SOX18 associated proteins, endothelial transcription factors and positive/negative control proteins. Right column: heatmap representation of Sm4 activity on SOX18-dependent protein-protein interactions, as tested at 100 μM. Interaction and disruption threshold is indicated in the scale bar by a black line. Levels of interaction and disruption above the threshold are demarked by ‘+’, and below the threshold by ‘−‘. Tagged proteins were expressed in the Leishmania tarentolae cell-free protein expression system. (D) Representative ALPHA-Screen concentration-response curve for SOX18 PPI disruption by Sm4. Data shown are mean ± s.e.m.
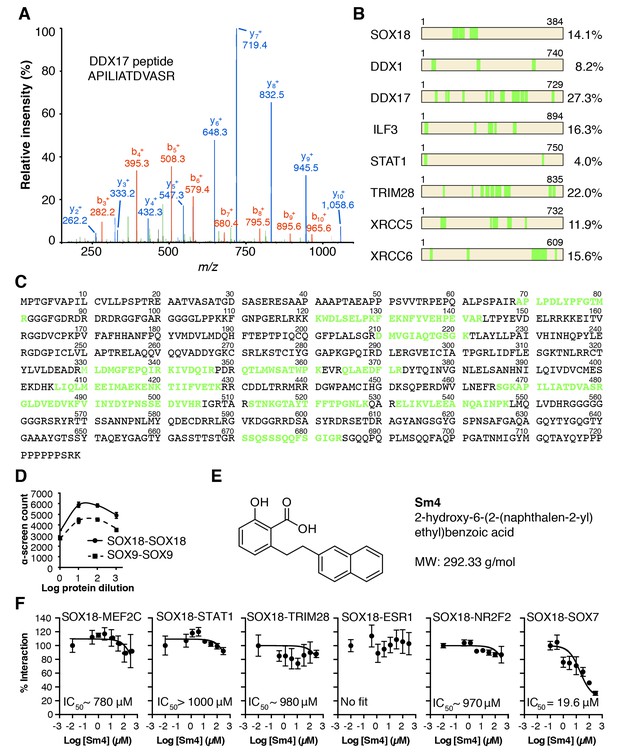
QC of SOX18 PPIs and effect of Sm4.
(A) Mass spectrometry spectrum for a representative double charged DDX17 peptide with the sequence KAPILIATDVASRG (Muscat ion score 51.6), identified from immunoprecipitation of cMyc-SOX18 with anti-cMyc antibody in HUVECs. (B) Coverage of identified peptides of SOX18 and interacting proteins selected from ChIP-MS. (C) Amino acid sequence of DDX17, with the identified ChIP-MS peptides indicated in green. (D) Typical ALPHA-Screen curve for protein dilution optimization, showing SOX9-SOX9 and SOX18-SOX18. The presence of a peak (hook effect) demonstrates an interaction and represents the ideal protein concentration for consecutive binding studies. Proteins were expressed in the Leishmania tarentolae cell-free protein expression system. (E) Molecular structure of SOX18 inhibitor Sm4.(F) ALPHA-Screen concentration-response curves for SOX18 PPI disruption by Sm4. Data shown are mean ± s.e.m.
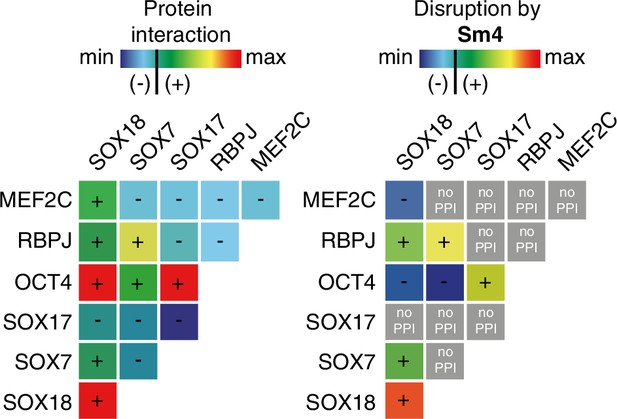
Differential disruption of SOXF PPI by Sm4.
The left panel shows a matrix of protein-protein interactions between SOXF, MEF2C and RBPJ and OCT4 as measured by ALPHAScreen. The right panel shows the effects of 50 µM Sm4 on PPIs (blue = no PPI/disruption, green/yellow = low PPI/disruption, orange/red = strong PPI/complete disruption, grey = PPI below threshold, Sm4 effect cannot be determined).
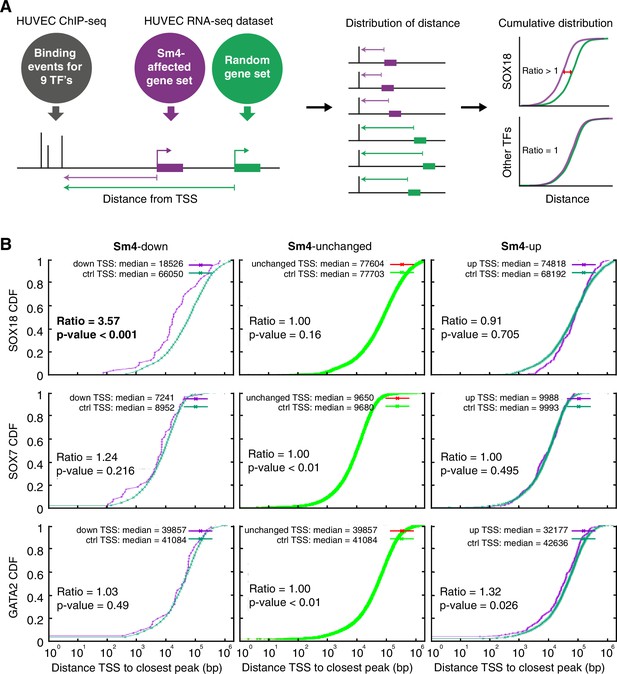
Sm4 selectively affects SOX18 transcriptional output in vitro.
(A) Schematic representation of the correlation analysis between genome-wide TF ChIP-seq data and Sm4 affected genes from transcriptomics data. The chromatin around the transcription start sites (TSS) of Sm4 affected genes (purple) was investigated for transcription factor binding peaks (grey), to calculate the ‘distance from TSS’ to closest binding site for a given transcription factor. This distance from TSS was used as a proxy for the likelihood of transcriptional regulation, and thus make an association between Sm4 affected genes and transcription factors. Included in the analysis where the ChIP-seq peaks of SOX18 and SOX7, and of all transcription factors available from the Encode consortium (GATA2, c-FOS, c-JUN, CTCF, EZH2, MAX and c-MYC), performed in HUVECs. A random group of genes was analysed as a control distribution as would be found by chance. (B) Sm4 affected genes were grouped into down-regulated (Sm4-down), unaffected (Sm4-unchanged) and up-regulated (Sm4-up). The plots show the cumulative distribution of the distance between the TSS of Sm4 affected genes (purple line, absolute fold change ≥2) and the closest genomic location of binding sites for SOX18, and control transcription factors SOX7 and GATA2. The median distance from the TSS of differentially expressed genes to the nearest binding event of a given transcription factor was compared to the median distance that is expected by chance from a random gene set (green line). Sm4 down regulated genes are significantly closer (bold) to the SOX18 peaks, but not to SOX7 or GATA2 peaks.
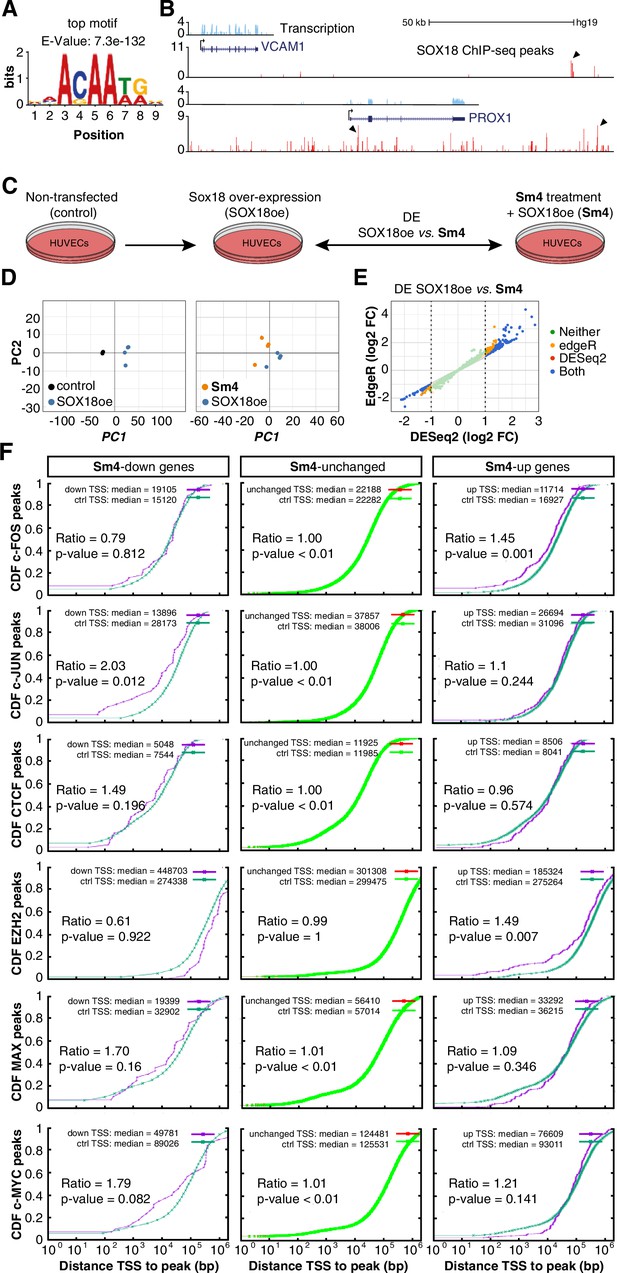
Transcriptome-wide analysis of Sm4 selectivity in vitro.
(A) Top motif identified from SOX18 ChIP-seq peaks (MEME software) performed in HUVECs. (B) UCSC browser view of representative ChIP-seq peaks (arrowheads) for known SOX18 target genes VCAM and PROX1. (C) Conditions for transcriptome-wide analysis of Sm4. Differential expression (DE) was calculated using DEseq2 in SOX18 overexpressing HUVECs, between vehicle DMSO (SOX18oe) and cells that received 25 μM Sm4 (Sm4) (D) Principal component analysis of quadruplicate RNA-seq samples. Replicates samples from same condition (control, SOX18oe, Sm4) cluster together. (E) Plot showing a comparison between DESeq2 and edgeR methods, marking significance of DE genes between SOX18oe and Sm4 conditions. Transcripts with a DESeq2 Log2 Fold Change ≥ 1 or ≤ −1 (dashed lines) were considered for further analysis. (F) The distance between Sm4 affected genes (purple) and the closest genomic location of binding sites a given transcription factor, plotted as cumulative distribution. The median distance from the TSS of differentially expressed genes to the nearest binding event of a transcription factor binding event was expressed as a ratio over the median distance that is expected by chance (random genes, green).

c-JUN motifs are enriched in SOX18 binding sites.
(A) HOMER motif analysis on SOX18 ChIP-seq peaks revealed an enrichment of the c-JUN motif 5’-TGAC/GTCA-3’. (B) ALPHA-Screen binding curve for SOX18-c-JUN and SOX18-SOX18 (positive control), demonstrating that c-JUN has the capacity to directly interact with SOX18 in vitro. Proteins were expressed in the Leishmania tarentolae cell-free protein expression system.

Sm4 does not interfere with SOX9 or SOX17 activity in vitro.
(A) Cell based reporter assay for SOX9 homodimer activity. COS-7 cell were transfected with Sox9 and Col2a1:luc reporter construct. Sox9 overexpression caused a >8 fold induction of Col2a1 activation. No change was observed at high concentration of Sm4. (B) Cell based reporter assay for SOX17 activity (Robinson et al., 2014). Bovine Aortic Endothelial Cells (BAECs) were transfected with pTK-β-gal (pTK) or ECE1-TK-β-gal (ECE1) reporter, measuring endogenous activity of SOX17 (ECE1-only). No change was observed at any of the tested concentration. Numbers on x-axis are [Sm4] in μM.
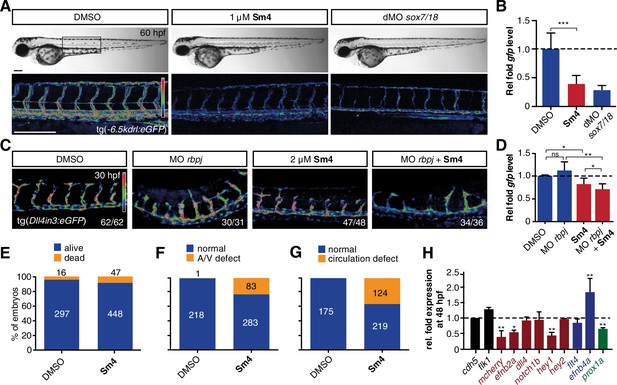
Sm4 blocks SoxF transcriptional activity in vivo.
(A) Lateral brightfield (top) and fluorescent (bottom) images of 60 hpf zebrafish larvae carrying the tg(−6.5kdrl:eGFP) SoxF reporter. Treatment was initiated at late stage (20 hpf) with either DMSO (negative control) or 1 μM Sm4, or larvae were injected with morpholinos against both sox7 and sox18 (dMO sox7/18). Fluorescence intensity is shown as heatmap. Scale bar 200 μm (B) qRT-PCR analysis on gfp transcripts levels in treated tg(−6.5kdrl:eGFP) larvae and sox7/18 morphants, showing reduction of activity on this transgene. (C) Lateral view of zebrafish larvae carrying the tg(Dll4in3:eGFP) SoxF/Notch reporter that harbors multiple binding sites for Rbpj and SoxF transcription factors. Larvae were injected with a morpholino against rbpj and/or treated with 2 μM Sm4 from 13 hpf. (D) qRT-PCR analysis on gfp transcripts in tg(Dll4in3:eGFP) larvae, showing repression of combined SoxF/Notch activity in the Sm4-treated larvae. (E) Quantitation of embryonic lethality in larvae, treated with Sm4 or DMSO control from early stage (16 hpf) until 72 hpf. (F) Penetrance of vascular phenotype (arteriovenous shunting) in 48 hpf larvae treated with 1.5 µM Sm4 from 16 hpf. (G) Penetrance of circulation defect in 48 hpf larvae treated with 1.5 µM Sm4 from 16 hpf. (H) qRT-PCR analysis of endogenous endothelial transcript levels at 48 hpf in larvae treated with 1.5 µM Sm4 at 16 hpf, relative to DMSO control (dotted line). Data shown are mean ± s.e.m. *p<0.05, **p<0.01, ***p<0.001.
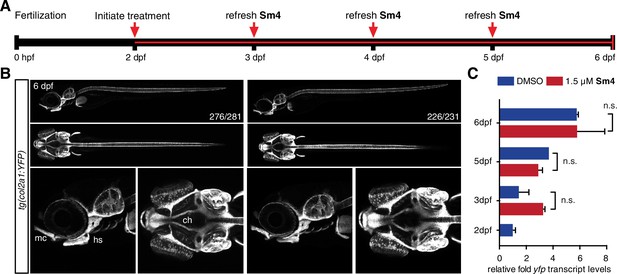
Sox9 activity is not perturbed by treatment in vivo.
(A) Timeline of treatment: Zebrafish larvae were treated continuously for four days during chondrogenesis. Medium was refreshed daily throughout the experiment to maintain Sm4 levels. (B) tg(col2a1:YFP) Sox9 reporter larvae marking cartilage (Mitchell et al., 2013). YFP levels were unaffected in presence of Sm4, and no changes in chondrogenesis were observed. mc: Meckel’s cartilage, ch: ceratohyal, hs: hyosymplectic. (C) qRT-PCR of yfp transcript levels in DMSO control and Sm4 treated larvae at a series of stages throughout chondrogenesis.

Sm4 interferes with SoxF activity in vivo.
(A) Timeline of Sm4 treatment in zebrafish larvae. Treatment for SOXF reporter gene studies was initiated at 20 hpf, while for the phenotypic studies treatment was initiated at precedes that for, to act during the right developmental window for arteriovenous specification. (B) Lateral view and transverse section of the trunk region of DMSO control and Sm4-treated tg(fli1:eGFP,−6.5kdrl:mCherry) larvae. Control DMSO larvae formed a distinctly separated dorsal aorta (DA) and posterior cardinal vein (PCV). In Sm4-treated larvae, the DA was constricted and/or fused to the PCV (arrowheads). Whole mount in situ hybridization against arterial marker efnb2a shows reduced expression and compromised formation of the DA and in Sm4-treated larvae at 48 hpf (arrows). Sections were DAPI stained (in blue). Scale bar brightfield: 0.5 mm, fluorescent and in situ 25 μm. (C) Concentration dependent effect of Sm4, showing quantitation for predominant phenotype at 72 hpf: mild (tail curvature), medium (dilation of the PCV) or severe (arteriovenous defect and/or circulation defect). Indicated timeframe refers to Sm4 treatment window and endpoint. (D) Quantitation of cardiac edema frequency in larvae treated with Sm4 (1.5 μM). (E) qRT-PCR analysis of Sox18 dependent −6.5kdrl:mCherry and endogenous endothelial transcript levels in Sm4-treated larvae relative to DMSO control (dotted line), showing effect on arterial and venous markers at 24 hpf. All expression levels were normalized to expression of endothelial marker cdh5. Data shown are mean ± s.e.m. *p<0.05, **p<0.01, ***p<0.001.
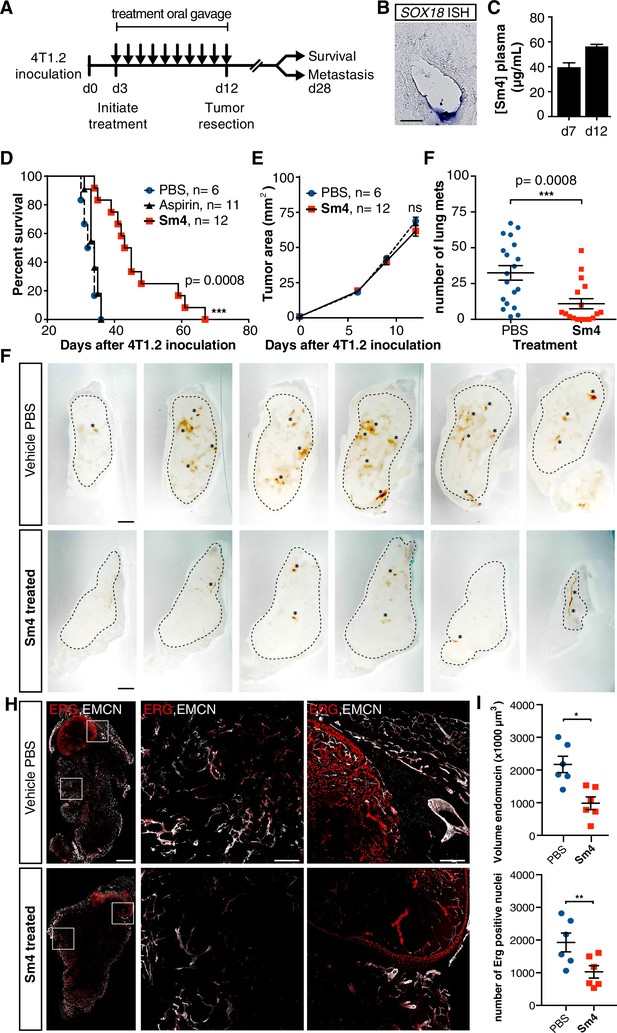
Metastasis and tumor vascularization is suppressed by Sm4 treatment.
(A) Timeline of mouse model for breast cancer metastasis. 4T1.2 tumor was inoculated at day 0, and resected at day 12. Sm4 (25 mg/kg/day), Aspirin (25 mg/kg/day) or vehicle control (PBS), was administered orally on a daily basis from day 3 to day 12. Independent experiments were carried out to assess survival and metastatic rate. (B) Blood plasma concentrations of Sm4 during the course of the treatment scheme (day 7 and day 12) indicate good systemic delivery of the drug. (C) Expression of SOX18 in the vasculature of the tumor as shown by in situ hybridization. Scale bar 100 μm. (D) Survival of the mice was monitored (n = 6–12 mice per group). Improved survival in Sm4-treated mice over both vehicle control and aspirin was analysed by Log-rank test (p<0.001). (E) No significant differences were found in tumor size at any stage. (F) Metastatic tumor nodules on the surface of the lungs were quantified at day 28, before any of the vehicle control or Sm4-treated animal had succumbed to the cancer burden. Data shown are mean ± s.e.m of 12–14 mice per group. (G) Vascular density was investigated on 300 μm sections of whole tumors. Bright field images show the overall morphology of the tumor (outlined by dashed line) and presence of red blood cells, marking the main blood vessels and haemorrhagic areas (asterisks). Scale bar 1 mm. (H) Double immunofluorescence staining for endothelial specific markers ERG and Endomucin (EMCN) reveals the vascular patterning and penetration in the intra- and peri- tumoral regions. Left: whole tumor section (scale bar 1 mm), middle and right: blow-up of boxed regions (scale bar 200 µm). (I) Quantitation of EMCN volume (blood vessel density) and ERG-positive nuclei (number of endothelial cells) of n = 6 tumours per condition. Each data point represents the average of 3–4 representative regions (boxed areas in panel H) per tumor. Mean ± s.e.m for both conditions are shown. *p<0.05, **p<0.01.
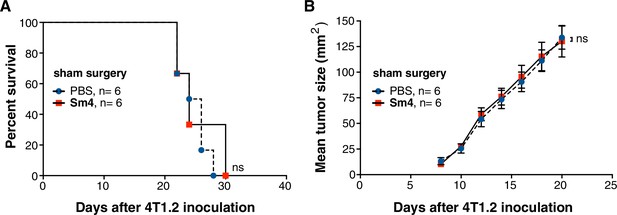
Sm4 efficacy is not a result of surgery-induced inflammation.
4T1.2 tumor was inoculated at day 0, and surgery was performed at day 12, without resecting the tumor (n = 6). (A) Survival (n = 6) was monitored in PBS vehicle control mice and Sm4-treated mice (25 mg/kg/day). No differences were observed (Log-rank test). (C) No significant differences were found in tumor size at any stage before or after sham surgery.
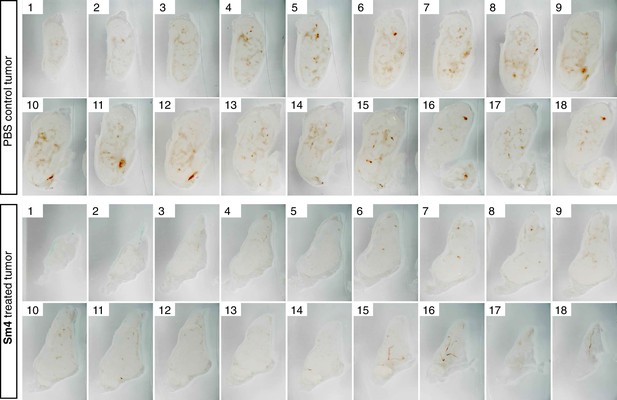
Penetrance of blood vessels into 4T1.2 tumors is impaired by Sm4.
Brightfield images of serial vibratome sections (300 μm) from a whole 4T1.2 mammary tumor for mice treated with PBS vehicle or Sm4. Main blood vessels and haemorrhagic areas are distinctive in red.
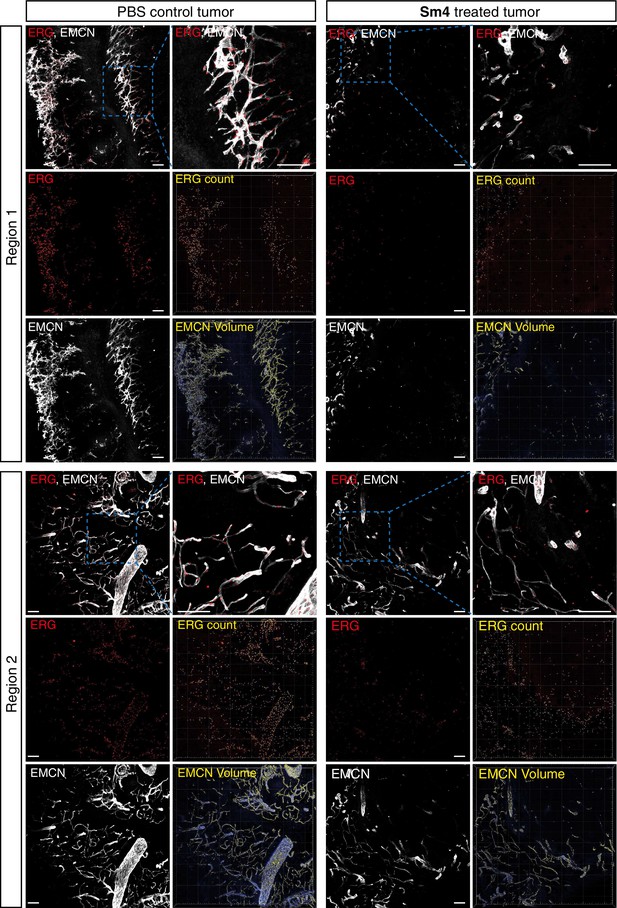
Sm4-treated mice have decreased tumor vascular density.
Immunofluorescent staining for ERG and Endomucin (EMCN) on tumor sections. Two representative regions for both vehicle PBS and Sm4 are shown. Detailed blow-up shows distinct nuclear staining for ERG, and membranous endothelial staining for EMCN. Quantitation of endothelial cells number and vascular volume was performed in Imaris on images with identical XYZ dimensions. Thresholds were chosen to accurately capture total EMCN+ vasculature and total ERG+ nuclei (ERG count and EMCN volume in yellow).
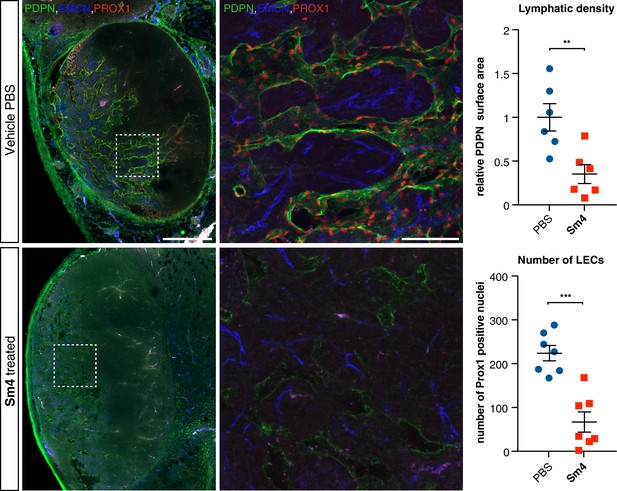
Sm4 treatment disrupts tumour-induced lymphangiogenesis.
Lymphatic vessels images of serial vibratome sections (200 μm) from a whole 4T1.2 mammary tumor for mice treated with PBS vehicle or Sm4 (25 mg/kg/day). Immunofluorescence for lymphatic specific markers PROX1 and Podoplanin (PDPN) and vascular EC marker Endomucin (EMCN) reveals the vascular patterning and penetration in the intra- and peri- tumoral regions. Whole tumor section for the control group (top panels), and for Sm4 treated group (bottom panels). Quantitation of PDPN+ lymphatic vascular area (density, top graph) and PROX1+ nuclei (number of lymphatic endothelial cells, bottom graph) of n ≥ 6 tumours per condition. Scale bar left: 0.5 mm, right: 0.1 mm. Mean ± s.e.m for both conditions are shown. **p<0.01, ***p<0.001.
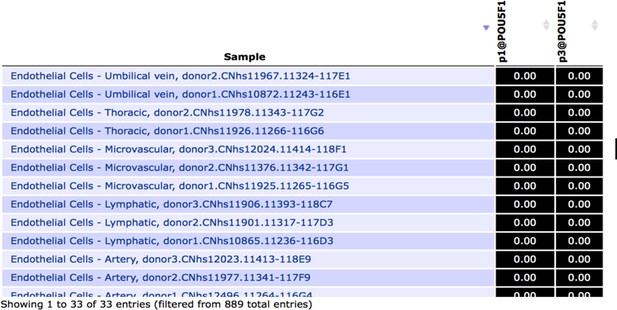
Snapshot of FANTOM5 database, showing (absence of) OCT4 transcript levels in arterial, venous and lymphatic endothelial cell types.
https://doi.org/10.7554/eLife.21221.018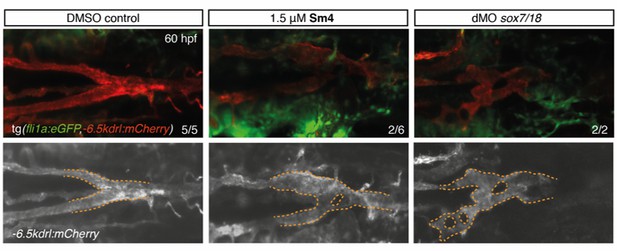
Sm4-treatment causes mild malformations to the lateral dorsal aorta (LDA), reminiscent of partial interference with Sox7 function.
Head circulation is unaffected by Sm4.
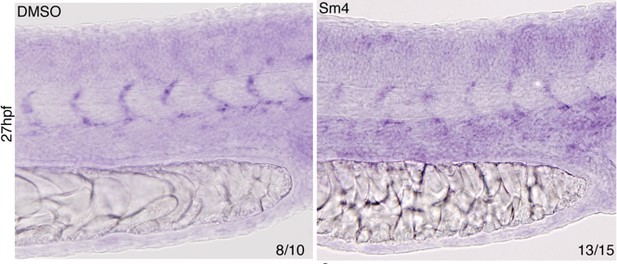
Effect of Sm4 on endogenous dll4 transcript in 27 hpf zebrafish larvae.
Both the dorsal aorta and intersomitic vessels (ISV) were labeled by dll4 ish probe. In presence of Sm4(1 μM) ISV show a mild decrease of signal intensity.
Additional files
-
Supplementary file 1
(A) GO term analysis (PANTHER) on top 5K SOX18 ChIP-seq peaks, reveals over-representation of biological processes, which are in agreement with known roles for SOX18 (e.g. blood vessel morphogenesis, angiogenesis, blood vessel development).
(B) Summary of sequencing statistics, listing the sample with the number of the replicate (#n). Percentage of mapped reads is consistently high across all samples (>87%). Mapping was performed with STAR aligner (Dobin et al., 2013). (C) Summary of endothelial specific TF expression levels and summary of distance from peak to TSS analysis on DE SOX18oe vs. Sm4 genes. A subtraction of SOX18, or cJUN peaks from all TF peaks was performed to reduce overlap bias (column #2 and #3). Sm4 down regulated genes are significantly closer to SOX18 and c-JUN ChIP-seq peaks.
- https://doi.org/10.7554/eLife.21221.017





