Epidermal Growth Factor Receptor neddylation is regulated by a desmosomal-COP9 (Constitutive Photomorphogenesis 9) signalosome complex
Figures
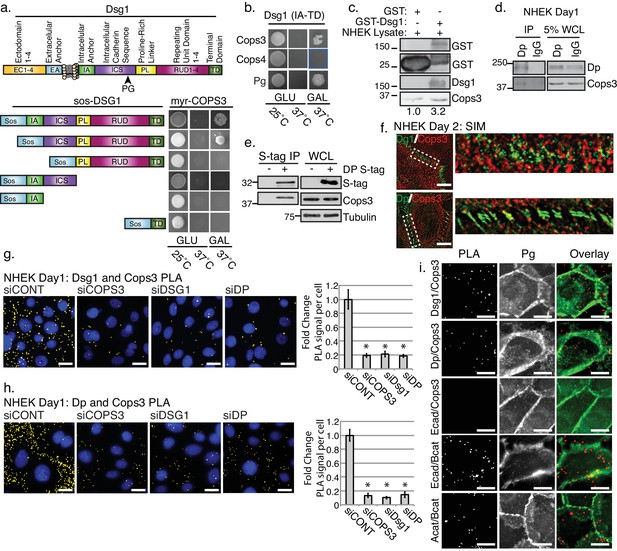
The third subunit of the COP9 signalosome (CSN), Cops3, interacts with desmosomal components Desmoglein1 (Dsg1) and Desmoplakin (Dp).
(a) Cytotrap yeast two hybrid (Y2H) indicating an interaction between Cops3 and the cytoplasmic tail of Dsg1. Growth at 25°C is permissive; growth on galactose (Gal) at 37°C confirms interaction. (b) Cops4 of the CSN does not interact with Dsg1. A known binding partner of Dsg1, Plakoglobin (Pg), was used as a positive control. (c) Purified GST-Dsg1 (includes the entire cytoplasmic domain), was bound to sepharose beads and incubated with normal human keratinocyte (NHEK) lysates. (d) An endogenous Dp immunoprecipitation (IP) was performed in NHEKs after 1 day of differentiation, indicating an interaction with Cops3. (e) A 32 kDa S-tagged C-terminal truncation of Dp (DP-S-tag) associates with Cops3 in squamous cell carcinoma line 9 (SCC9) lysates. (f) Structured Illumination Microscopy (SIM) of Cops3 (red) and Dsg1 or Dp (green) in NHEKs after 2 days of differentiation, shows close proximity between Cops3 and desmosome components at junctions. (g and h) Proximity ligation assay (PLA) in NHEK cultures with siRNA-targeting COPS3, DSG1, DP, or scramble sequence (siCONT) after 1 day of differentiation. Quantification is displayed to the right of the image panels. Cops3/Dsg1= *p<0.001, and Cops3/Dp= *p<0.00005, Student’s t test; mean ±SEM. (i) PLA in NHEK cultures after 1 day of differentiation with adherens junction components (Ecadherin, E-cad, Beta catenin, B-cat, Alpha catenin, A-cat) and co-stained with Plakoglobin to mark cell borders. Proximal proteins within 40–100 nm appear as red but have been pseudo-colored yellow in (g) and (h) and have been dilated in (i) for ease of visualization in Fiji. All experiments are representative of 3 or more independent repeats. Scale bars = 20 μm.
-
Figure 1—source data 1
PLA analysis of Dsg1 and Cops3 in NHEKs differentiated for 1 day.
Quantification of raw data of PLA signal per cell for NHEKs cultures with siRNA-targeting COPS3, DSG1, DP, or scramble sequence (siCONT) after 1 day of differentiation. Quantification performed in FIJI of proximal proteins (see sample size and statistical analysis). *p<0.001 (Student’s t test); mean ±SEM (Figure 1g).
- https://doi.org/10.7554/eLife.22599.008
-
Figure 1—source data 2
PLA analysis of Dp and Cops3 in NHEKs differentiated for 1 day.
Quantification of raw data of PLA signal per cell for NHEKs cultures with siRNA-targeting COPS3, DSG1, DP, or scramble sequence (siCONT) after 1 day of differentiation. Quantification performed in FIJI of proximal proteins (see sample size and statistical analysis). *p<0.00005 (Student’s t test); mean ±SEM (Figure 1h).
- https://doi.org/10.7554/eLife.22599.009
-
Figure 1—source data 3
PLA analysis of desmosomal and adherens junctions components in NHEKs differentiated for 1 day.
Quantification of raw data of PLA signal per cell for NHEKs cultures with siRNA-targeting Cops3 (Figure 1—figure supplement 4a and b). Figure 1—figure supplement 4c displays the percent of PLA signal at cell junctions facilitated by co-staining of cell-cell junctions with Pg. Quantification performed in FIJI of proximal proteins (see sample size and statistical analysis).
- https://doi.org/10.7554/eLife.22599.010
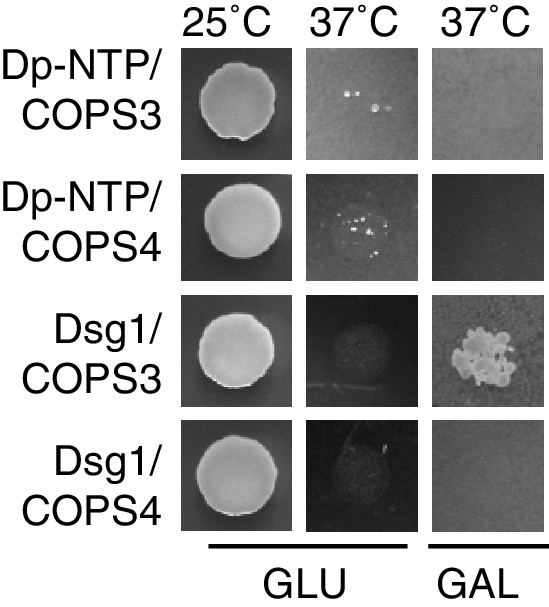
Cytotrap Y2H indicating no interaction between Cops3 and the N-terminal portion of Dp (amino acids 1–584).
Growth at 25°C is permissive; growth on galactose (Gal) at 37°C confirms interaction, while lack of growth on glucose (Glu) plates at 37°C excludes the presence of any false positives (ie. temperature-sensitive revertants).
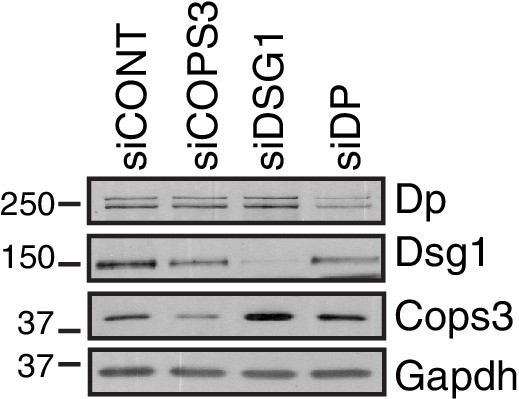
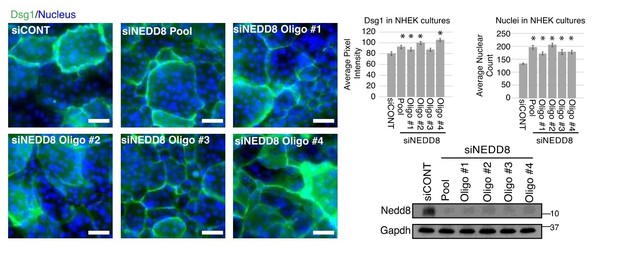
Western blots indicating knockdown efficiency of NHEKs used in proximity ligation assay (PLA) of Dsg1 and Cops3.
https://doi.org/10.7554/eLife.22599.005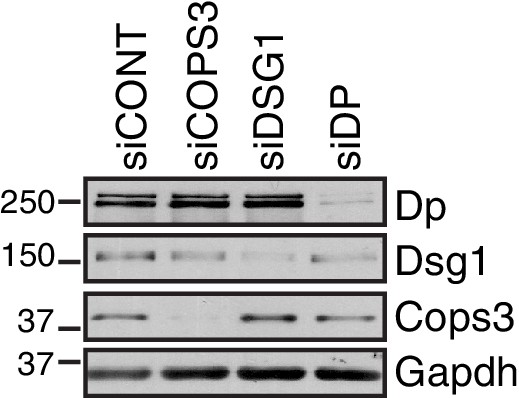
Western blots indicating knockdown efficiency of NHEKs used in proximity ligation assay (PLA) of Dp and Cops3.
https://doi.org/10.7554/eLife.22599.006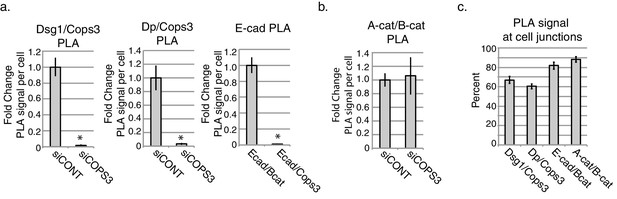
Quantification of PLA analysis in NHEKS after 1 day of differentiation.
Antibody pairs are (a) Dsg1/Cops3, Dp/Cops3 and E-cad/B-cat, and Ecad/Cops3. *p<0.000005, Student’s t test; mean ±SEM. (b) Antibody pairs are adherens junction components Alpha Catenin (A-cat) and Beta catenin (B-cat). Close association of these adherens junction components are unaffected upon the loss of Cops3. (c) Quantification of PLA analysis to distinguish percent PLA signal at Pg-positive staining borders. Experiment is representative of 3 or more independent repeats.
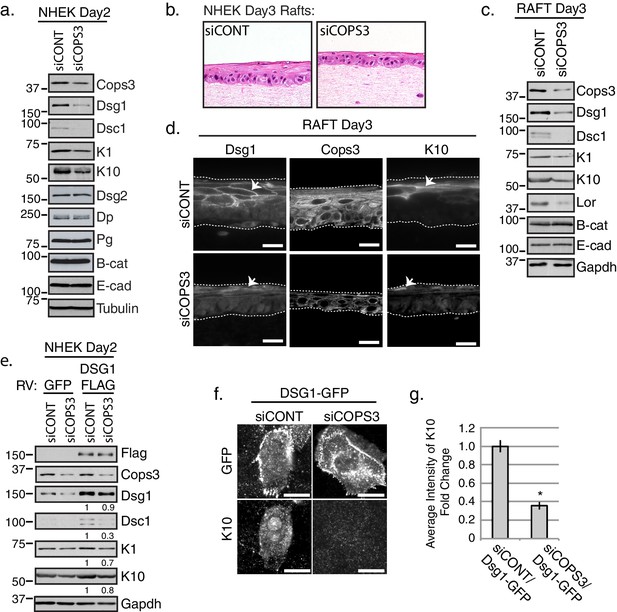
Keratinocyte differentiation requires Cops3.
(a) RNAi-mediated silencing of COPS3 in NHEK cultures differentiated for 2 days in 1.2mM Ca++results in a decrease in differentiation markers: Dsg1, Desmocollin1 (Dsc1), Keratin 10 (K10) and Keratin1 (K1). No change in protein expression for Desmoglein2 (Dsg2), Dp, Pg, Beta-catenin (B-cat), Ecadherin (E-cad) was seen between siCONT and siCOPS3 cell lysates. Tubulin was utilized as a loading control. (b) 3D epidermal raft cultures were generated with cells silenced by siRNA-targeting COPS3 or scramble sequence (siCONT) and harvested 3 days after lifting to an air-medium interface. H & E staining and biochemical analysis of raft cultures display signs of impaired differentiation. (c) Biochemical analysis of 3D raft lysates corresponds to morphology changes with an observed decrease in Dsg1, Dsc1, K10, K1, and Loricrin (Lor) with no changes in B-cat and E-cad. (d) Immunofluorescence of siCOPS3 raft cultures displays a decrease in Dsg1 and K10 (scale bar = 20 μm). (e) Cells silenced for Cops3 were transduced with GFP or Dsg1-FLAG retrovirus (RV). Ectopic expression of Dsg1 was unable to fully compensate for the differentiation defect found in Cops3 deficient cells, which was analyzed by differentiation markers (Dsc1, K1, K10). Densitometry quantifications of differentiation markers are represented as fold change relative to Gapdh and indicated below the blot. (f) NHEKs transiently transfected with the Dsg1-GFP construct and processed after 1 day of differentiation do not express K10 upon the silencing of COPS3 (scale bar = 20 μm). (g) Quantification of average fluorescence intensity of K10 in Dsg1-GFP expressing cells. *p<0.00005 (Student’s t test); mean ±SEM. All experiments are representative of 3 or more independent repeats.
-
Figure 2—source data 1
Cells transiently transfected with Dsg1-GFP do not express K10 upon the silencing of COPS3.
Quantification of raw data of average fluorescence intensity in Dsg1-GFP expressing cells. Quantification performed in FIJI (see sample size and statistical analysis). *p<0.00005 (Student’s t test); mean ±SEM (Figure 2g).
- https://doi.org/10.7554/eLife.22599.013
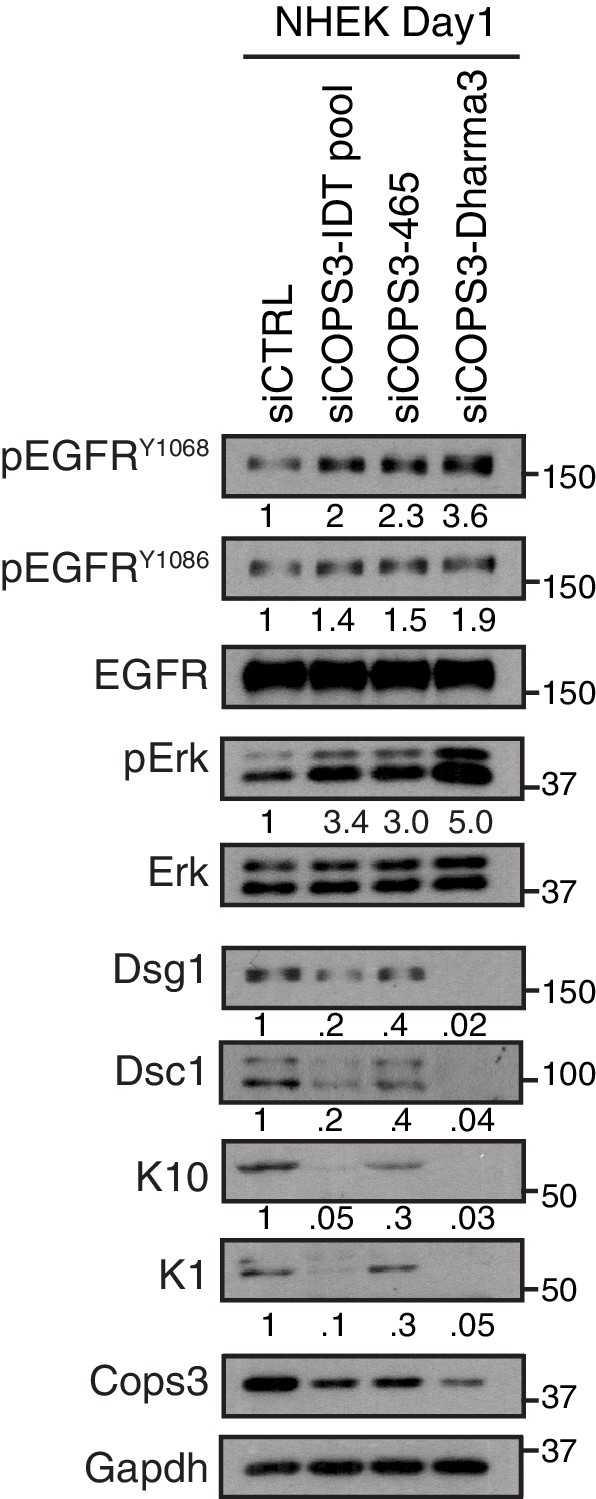
Multiple siRNA targets of COPS3 displayed keratinocyte differentiation defect and an increase in EGFR signaling in keratinocytes differentiated for 1 day in 1.2 mM Ca++.
https://doi.org/10.7554/eLife.22599.012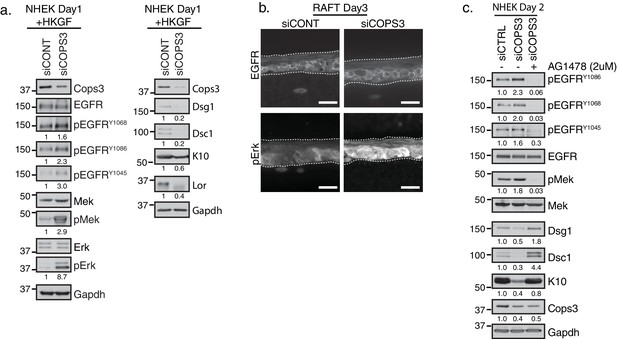
Silencing of Cops3 promotes Epidermal Growth Factor Receptor (EGFR) signaling in NHEKs.
(a) RNAi-mediated silencing of COPS3 in NHEKs differentiated for 2 days in 1.2 mM Ca++ with Human Keratinocyte Growth Factors (HKGF) display a decrease in differentiation markers (right panel) and an increase in phosphorylation of EGFR, along with its downstream effectors (Mek and Erk), indicating an overall increase in MAPK signaling (left panel). For quantification of phosphorylated proteins, each band was normalized to the amount of total protein then compared relative to the control culture (siCONT) and indicated below each western blot. (b) Immunofluorescence of siCOPS3 raft cultures display similar levels of total EGFR and an increase in pErk signal (scale bar = 20 µm). (c) Western blot analysis of NHEKs treated with siRNA targeting scramble (siCONT) and COPS3 (siCOPS3) and incubated with an EGFR specific inhibitor, AG1478, at 2 μM. EGFR inhibition restored the expression of the differentiation markers, Dsg1, Dsc1, and K10, indicating that Cops3 operates upstream of EGFR signaling in the epidermal differentiation program. Densitometry quantification is represented as fold change relative to Gapdh and indicated below the blot. All experiments are representative of 3 or more independent repeats.
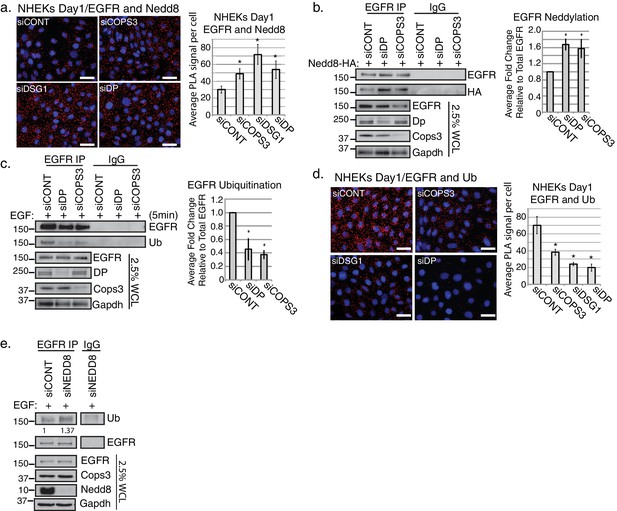
Loss of desmosomal components, Cops3 or Nedd8 affects the biochemical status of EGFR.
(a) PLA analysis using Nedd8 and EGFR antibodies in NHEKs treated with siRNA targeting DP, DSG1, COPS3 or scramble (siCONT) (scale bar = 20 µm). Quantification of PLA signals displayed to the right of the images. *p<0.05 (Student’s t test); mean ±SEM. (b) EGFR immunoprecipitations (IPs) in SCC9 cells harboring the Nedd8-HA construct indicate an increase in EGFR neddylation upon silencing of Cops3 or Dp. Densitometry quantification of 4 independent replicates is shown to the right of the representative blot. *p<0.05 (Student’s t test); mean ±SEM. (c) Cells treated with siRNA directed towards COPS3 or DP display a decrease in EGFR ubiquitination. EGFR IPs in SCC9 cells treated with 50 ng/ml EGF for 5 min to induce internalization of the receptor. Densitometry quantification of 4 independent replicates is shown to the right of the representative blot. *p<0.01 (Student’s t test); mean ±SEM. (d) PLA analysis using Ubiquitin and EGFR antibodies in NHEKs treated with siRNA targeting DP, DSG1, COPS3 or scramble (siCONT) (scale bar = 20 µm). Quantification of PLA signals displayed to the right of the images. *p<0.05 (Student’s t test); mean ±SEM. (e) EGFR immunoprecipitations (IPs) in SCC9 cells indicate EGFR ubiquitination upon silencing of Nedd8 and treated with 50 ng/ml EGF ligand for 5 min. Western blot displayed is a representative blot of 3 independent repeats. All experiments are representative of 3 or more independent repeats.
-
Figure 4—source data 1
PLA analysis of Nedd8/EGFR antibodies and Ubiqutin/EGFR antibodies in NHEKs.
Quantification of raw data of PLA signal per cell treated with siRNA targeting DP, DSG1, COPS3 or scramble (siCONT). Quantification performed in FIJI of proximal proteins (see sample size and statistical analysis). Nedd8/EGFR: *p<0.05 (Student’s t test); mean ±SEM (Figure 4a). Ubiquitin/EGFR: *p<0.05 (Student’s t test); mean ±SEM (Figure 4d).
- https://doi.org/10.7554/eLife.22599.018
-
Figure 4—source data 2
Analysis of Nedd8-EGFR and Ubiquitin-EGFR through EGFR immunoprecipitations (IPs) in SCC9 cells.
Densitometry quantification of raw data of 4 biological replicates. Nedd8 and Ubiquitin quantifications were normalized to the total amount of EGFR immunoprecipitated from each biological replicate. Quantification performed in FIJI of proximal proteins (see sample size and statistical analysis). Nedd8-EGFR: *p<0.05 (Student’s t test); mean ±SEM (Figure 4b). Ubiquitin-EGFR: *p<0.01 (Student’s t test); mean ±SEM (Figure 4c).
- https://doi.org/10.7554/eLife.22599.019
-
Figure 4—source data 3
PLA analysis of EGFR/Nedd8 and EGFR/Ub upon the loss of Nedd8.
Quantification of raw data of PLA signal per cell for NHEKs cultures with siRNA-targeting NEDD8 and COPS3 after 1 day of differentiation. Quantification performed in FIJI of proximal proteins (see sample size and statistical analysis). *p<0.00005 (Student’s t test); mean ±SEM (Figure 4—figure supplement 2).
- https://doi.org/10.7554/eLife.22599.020

Western blots indicating knockdown efficiency of NHEKs used in proximity ligation assay (PLA) of EGFR/Nedd8 and EGFR/Ub (Fig.
https://doi.org/10.7554/eLife.22599.016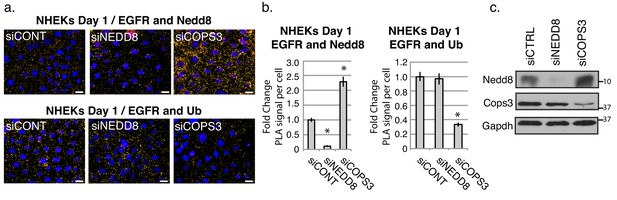
EGFR is ubiquitinated upon loss of Nedd8 by PLA analysis in NHEKs differentiated for 1 day in high calcium.
(a) PLA analysis of NHEKs differentiated after 1 day with siRNA targeting NEDD8 and COPS3. Antibody pairings: AB-12 anti-EGFRMOUSE/Ab81264 anti-Nedd8RABBIT(Top panel) and FK1 anti-UbMOUSE/D38B1 anti-EGFRRABBIT(Bottom panel). Proximal proteins within 40–100 nm are indicated by a yellow dot (signal appears as red and pseudo-colored yellow for ease of visualization in ImageJ) (b) Quantification of PLA signals per cell. *p<0.00005 (Student’s t test); mean ±SEM. (c) Western blot analysis indicates knockdown efficiency of siRNAs directed towards Nedd8 and Cops3. Experiment is representative of 3 or more independent repeats. Scale bar = 20 µm.
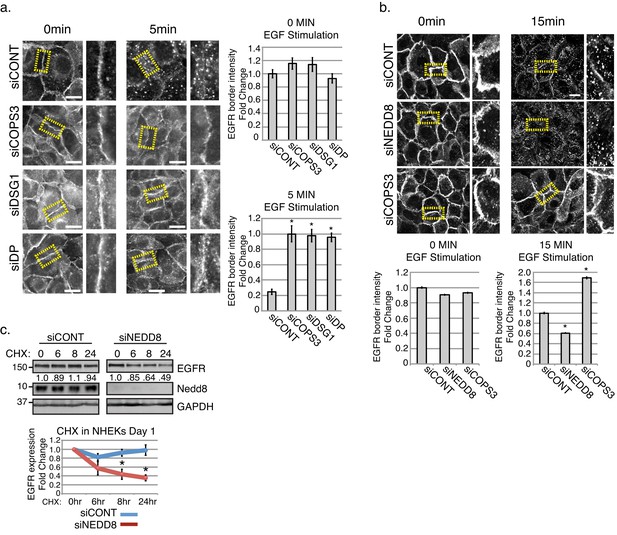
Stability of EGFR and the receptor’s internalization are affected upon the loss of desmosomal components, Cops3 or Nedd8.
(a) NHEKs were treated in serum free media (1.2 mM Ca++), then treated with EGF (50 ng/ml) at indicated time points to assess the amount of internalized EGFR (scale bars = 20 μm). Quantification of EGFR fluorescence border intensity from 3 independent replicates is displayed to the right of the images. *p<0.01 (Student’s t test); mean ±SEM. (b) NHEKs were treated in serum free media (1.2 mM Ca++), then treated with EGF (100 ng/ml) at indicated time points to assess the amount of EGFR remaining at cell borders upon the loss of Cops3 and Nedd8 (scale bars = 20 μm). Quantification of EGFR fluorescence border intensity from 3 independent replicates is displayed below the images. *p<0.05 (Student’s t test); mean ±SEM. (c) NHEKs were differentiated in high calcium for 1 day, and then treated with cycloheximide (CHX 0.02 mg/ml) for indicated times. Quantification of total EGFR is from 3 independent replicates and is normalized to GAPDH. *p<0.02 (Student’s t test); mean ±SEM. All experiments are representative of 3 or more independent repeats.
-
Figure 5—source data 1
EGFR localization upon stimulation in cells treated with siRNA targeting DP, DSG1, COPS3, or scramble (siCONT).
Quantification of raw data of EGFR fluorescence border intensity from 3 biological replicates. Quantification performed in FIJI (see sample size and statistical analysis). *p<0.01 (Student’s t test); mean ±SEM (Figure 5a).
- https://doi.org/10.7554/eLife.22599.023
-
Figure 5—source data 2
EGFR localization upon stimulation in cells treated with siRNA targeting NEDD8, COPS3, or scramble (siCONT).
Quantification of raw data of EGFR fluorescence border intensity from 3 biological replicates. Quantification performed in FIJI (see sample size and statistical analysis). *p<0.01 (Student’s t test); mean ±SEM (Figure 5b).
- https://doi.org/10.7554/eLife.22599.024
-
Figure 5—source data 3
Cycloheximide (CHX) treatment of NHEKS upon the loss of Nedd8.
Densitometry quantification of raw data of 3 biological replicates. Total EGFR protein expression was normalized to loading control GAPDH for each indicated time point. Quantification performed in FIJI (see sample size and statistical analysis). 8 hr CHX treatment = *p<0.02; and 24 hr CHX treatment = *p<0.01. Student’s t test; mean ±SEM (Figure 5c).
- https://doi.org/10.7554/eLife.22599.025
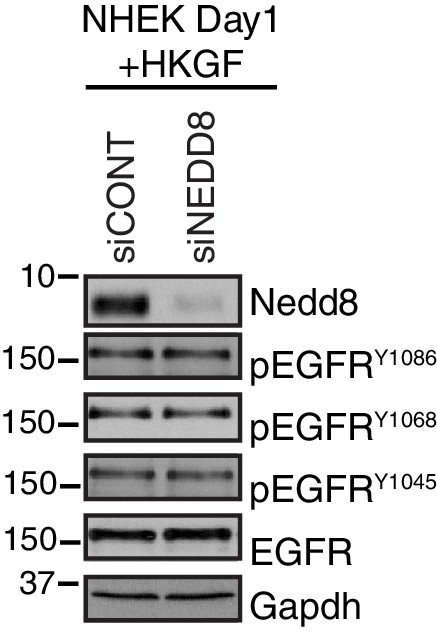
Western blot analysis of NHEKs differentiated for 1 day in 1.2 mM Ca++ with Human Keratinocyte Growth Factor (HKGF) supplement to visualize EGFR signaling at different phosphorylation sites upon the loss of Nedd8.
https://doi.org/10.7554/eLife.22599.022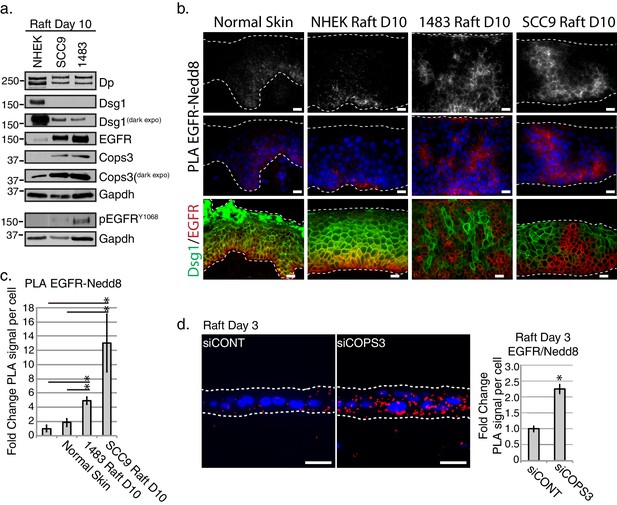
EGFR neddylation is increased in 3D Head and Neck Squamous Cell Carcinoma (HNSCC) and Cops3 knockdown organotypic models.
(a) Biochemical analyses of HNSCC 3D organotypic raft cultures generated using SCC9 and 1483 cell lines displayed a decrease in Dsg1 and Dp, and an increase in EGFR and Cops3. (b) PLA analysis of EGFRRABBIT and Nedd8MOUSE in normal skin, and 3D organotypic raft cultures generated using the NHEKs, SCC9s, and 1483 HNSCC cell lines (scale bar = 20 µm). Top panel displays PLA signal alone with an apparent cell border signal, and middle panel represents the PLA signal with Dapi overlay. Bottom panel displays fluorescent staining of DsgGREEN and EGFRRED in normal skin, and NHEK, SCC9 or HNSCC rafts. (c) Quantification of PLA signals in 3D cancer models. *p<0.05 (Student’s t test); mean ±SEM. (d) PLA analysis of 3D organotypic raft cultures generated with NHEKs treated with siRNA scramble sequence (siCONT) or siRNA directed towards Cops3 (siCOPS3), indicating an increase in EGFR-Nedd8 PLA signal (scale bar = 20 µm). Quantification of PLA signals displayed to the right of the images. *p<0.05 (Student’s t test); mean ±SEM. All experiments are representative of 3 or more independent replicates.
-
Figure 6—source data 1
PLA analysis Nedd8/EGFR in 3D cultures.
Quantification of raw data of PLA signal per cell of Nedd8/EGFR in normal skin, and 3D organotypic raft cultures generated using the NHEKs, SCC9s, and 1483 HNSCC cell lines. Quantification performed in FIJI of proximal proteins (see sample size and statistical analysis). *p<0.05 (Student’s t test); mean ±SEM (Figure 6c).
- https://doi.org/10.7554/eLife.22599.027
-
Figure 6—source data 2
PLA analysis of Nedd8/EGFR 3D organotypic deficient in Cops3.
Quantification of raw data of PLA signals of 3D raft cultures generated with NHEKs treated with siRNA scramble sequence (siCONT) or siRNA directed towards Cops3 (siCOPS3). Quantification performed in FIJI of proximal proteins (see sample size and statistical analysis). *p<0.05 (Student’s t test); mean ±SEM (Figure 6d).
- https://doi.org/10.7554/eLife.22599.028
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.22599.029