A subset of ipRGCs regulates both maturation of the circadian clock and segregation of retinogeniculate projections in mice
Figures
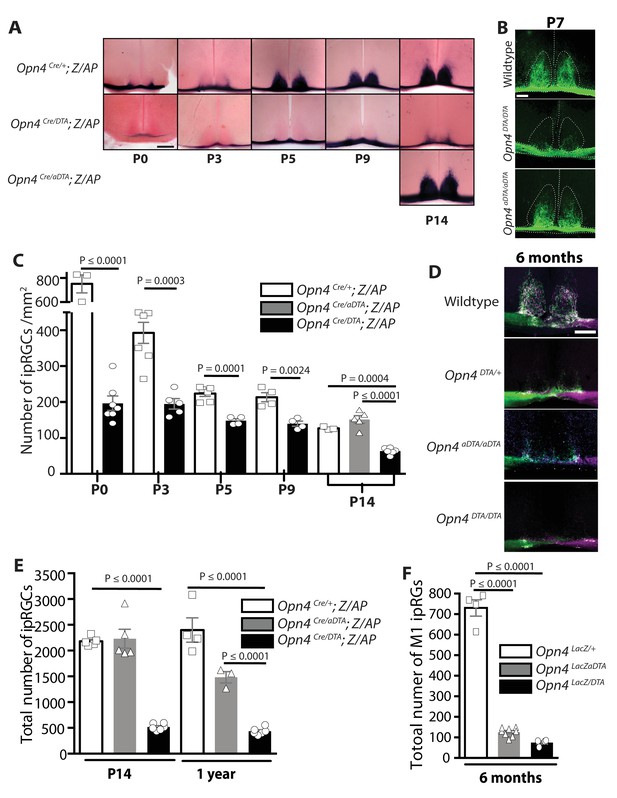
Developmental ablation of ipRGCs in the mouse retina.
(A) Developmental time course of ipRGC innervation of the SCN, visualized by AP staining, in Opn4Cre/+; Z/AP and Opn4Cre/DTA; Z/AP mouse. For comparison, SCN staining from Opn4Cre/aDTA; Z/AP mice at P14 are also shown. Scale bar = 200 μm. (B) SCN innervation in P7 WT, Opn4DTA/DTA, and Opn4aDTA/aDTA mice revealed by CTB injections into the eyes. Scale bar = 100 μm. (C) Developmental time course of ipRGC (all subtypes) cell density visualized by AP staining of retina from Opn4Cre/+ Z/AP (control) and Opn4Cre/DTA; Z/AP mice at P0 (control n = 3, DTA n = 7), P3 (control n = 7, DTA n = 5), P5 (control n = 6, DTA n = 4), P9 (control n = 4, DTA n = 4), and P14 (control n = 3, DTA n = 6, aDTA n = 5). Cell counts from P14 retinas of Opn4Cre/aDTA; Z/AP mice are also shown for comparison. Using a two-way ANOVA, we found a strongly significant effect of genotype. A t-test for P0, P3, P5, and P9 time points, and a one-way ANOVA with Bonferroni's post-hoc analysis for P14 revealed a significant cell loss at each time point. (D) SCN innervation revealed by CTB injections into the eyes of 6-month-old WT, Opn4DTA/+, Opn4DTA/DTA, and Opn4aDTA/aDTA mice. Scale bar = 200 μm. (E) Total cell counts of ipRGCs (all subtypes) revealed by alkaline phosphatase staining at P14 and 1 year of age in Opn4Cre/+; Z/AP (control; P14: n = 5, 1 year: n = 4), Opn4Cre/aDTA; Z/AP (P14: n = 5; 1 year: n = 3), and Opn4Cre/DTA; Z/AP (P14: n = 6; 1 year: n = 6). Two-way ANOVA, Bonferroni's multiple comparisons test and adjusted p values. (F) Total cell counts of M1 ipRGCs, identified by x-Gal staining of retinas from 6 month old Opn4LacZ/+ (control; n = 4), Opn4LacZ/aDTA, and Opn4LacZ/DTA mice (n = 4). One-way ANOVA, Bonferroni's multiple comparisons test and adjusted p values. Error bars represent s.e.m. for all graphs. See also Figure 1—figure supplement 1.
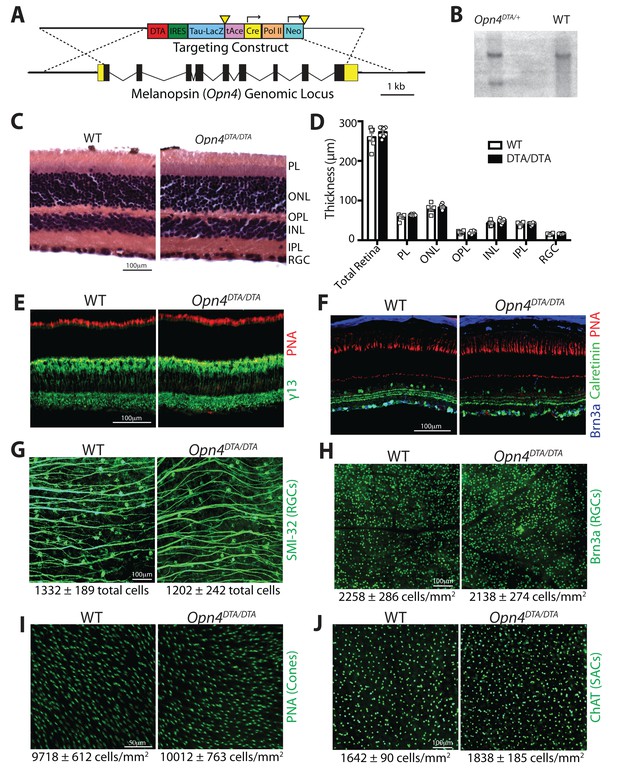
Generation and characterization of mice with an Opn4DTA allele.
(A) Targeting construct for inserting the coding sequence of DTA into the melanopsin (Opn4) locus. (B) Southern blot analysis confirming homologous recombination of the targeting construct at Opn4 locus in ES cells. (C) Hematoxylin and eosin staining on retinal sections from WT and Opn4DTA/DTA mice. (D) Quantification of the retinal layer thickness based on hematoxylin and eosin staining. No significant differences were found by a Student’s t-test. (E) Staining with fluorescently conjugated peanut agglutinin to label cones (red), and immunohistochemistry for caleretinin-positive amacrine and ganglion cells (green), and Brn3a-positive ganglion cells (blue) on retinal sections from WT and Opn4DTA/DTA mice. Scale bar = 100 µm. (F) Staining with fluorescently conjugated peanut agglutinin to label cones (red) and immunohistochemistry for γ−13 to label ON-bipolar cells (green) on retinal sections from WT and Opn4DTA/DTA mice. Scale bar = 100 µm. (G and H) We used two markers that label distinct populations of conventional RGCs: Brn3a is a marker for ~80% of conventional RGCs and it does not colocalize with melanopsin. SMI-32 labels alpha-like RGCs, a subset of which are M4 ipRGCs. Both RGC labeling methods showed no significant difference between Opn4DTA/DTA and WT. These data indicate that conventional RGCs and even a subtype of ipRGCs that expresses low levels of melanopsin are not ablated in Opn4DTA/DTA mice. Scale bar = 100 µm. (I) Density of cones, identified by staining with fluorescently conjugated peanut agglutinin, was not significantly different between Opn4DTA/DTA and WT. Scale bar = 50 µm. (J) We used ChAT staining to label Starburst amacrine cells, which are important for retinal waves. We found that the density of ChAT positive amacrine cells was similar between WT and Opn4DTA/DTA mice. Scale bar = 100 µm. (G–J) No significant differences were found by a Student’s t-test.
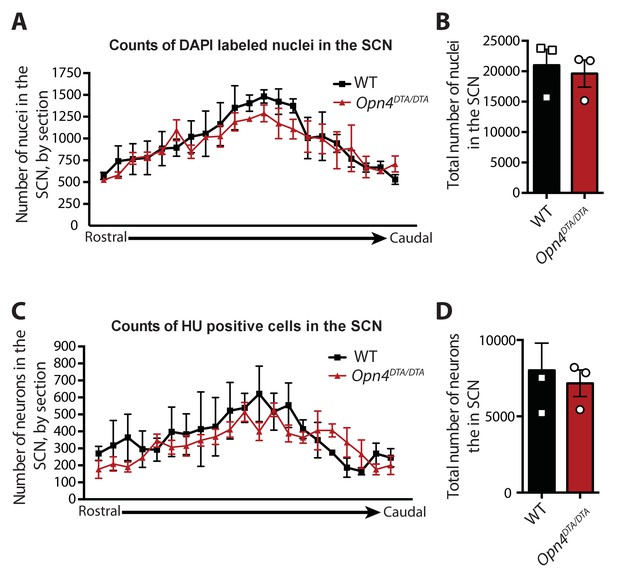
The number of cells in the SCN is unaffected in Opn4DTA/DTA mice.
(A) Number of DAPI-labeled nuclei in the SCN of WT and Opn4DTA/DTA mice graphed rostral to caudal based on 25 μm coronal sections. (B) Total number of DAPI-labeled nuclei in the SCN of WT and Opn4DTA/DTA mice. (A) Number of neurons (identified by anti-Hu immunohistochemistry) in the SCN of WT and Opn4DTA/DTA mice by 25 μm coronal section graphed rostral to caudal based on 25 μm coronal sections. (B) Total number of neurons in the SCN of WT and Opn4DTA/DTA mice. (A and C) No significant differences by two-way ANOVA, Bonferroni's multiple comparisons test and adjusted p values. (B and D) No significant differences were found by a Student’s t-test. Error bars for all graphs represent s.e.m.
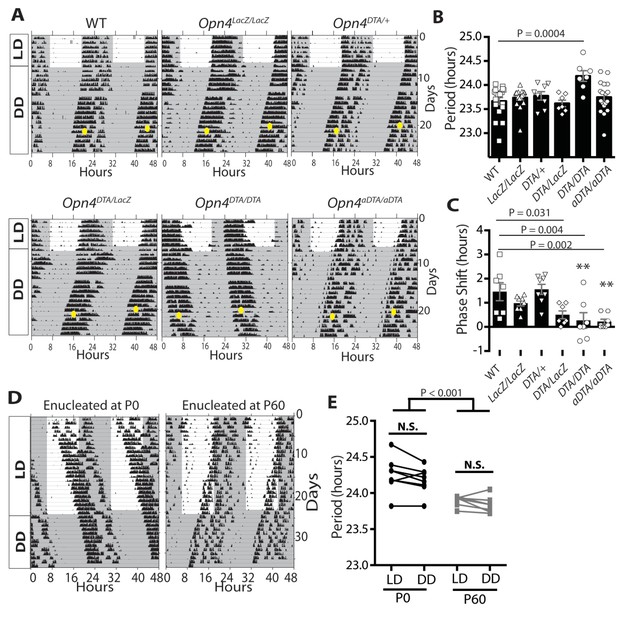
Developmental ablation of ipRGCs results in a lengthened circadian period.
(A) Representative actograms of wild-type, Opn4LacZ/LacZ, Opn4DTA/+, Opn4DTA/LacZ, Opn4DTA/DTA and Opn4aDTA/aDTA mice under 12:12-LD cycle and constant darkness (DD). White background indicates light and grey background indicates darkness. (B) The circadian period length of all tested genotypes (wild-type: n = 18, Opn4LacZ/LacZ: n = 17, Opn4DTA/+: n = 8, Opn4DTA/LacZ: n = 7, Opn4DTA/DTA: n = 7, and Opn4aDTA/aDTA: n = 18). (C) For phase-shifting experiments, a subset of wild-type, Opn4LacZ/LacZ, Opn4aDTA/aDTA were assessed, and the total mice analyzed was as follows: wild-type (n = 7), Opn4LacZ/LacZ (n = 8), Opn4DTA/+ (n = 8), Opn4DTA/LacZ (n = 7), Opn4DTA/DTA (n = 7), and Opn4aDTA/aDTA (n = 8). As expected from substantial ipRGC ablation, phase shifting was significantly reduced in Opn4DTA/LacZ, Opn4DTA/DTA and Opn4aDTA/aDTA mice. (D) Representative actograms of wild-type mice enucleated at either P0 (n = 8) or P60 (n = 7), under 12:12-LD and DD. (E) The circadian period length for both enucleation groups. Only mice enucleated at P0 exhibited a lengthened circadian period (B and C) One-way ANOVA with Bonferroni's multiple comparisons test and adjusted p values. (E) Two-way ANOVA, Bonferroni's multiple comparisons test and adjusted p values. Error bars represent s.e.m. for all graphs. See also Figure 2—figure supplements 1–2.
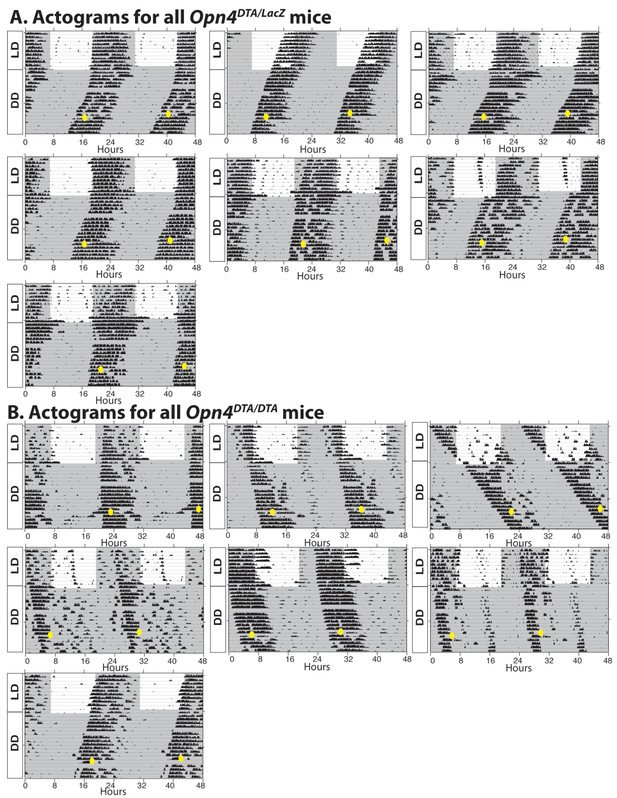
Additional actograms for Opn4DTA/LacZ and Opn4DTA/DTA mice.
(A) Actograms of all tested Opn4DTA/LacZ. (B) Actograms of all tested Opn4DTA/DTA mice.
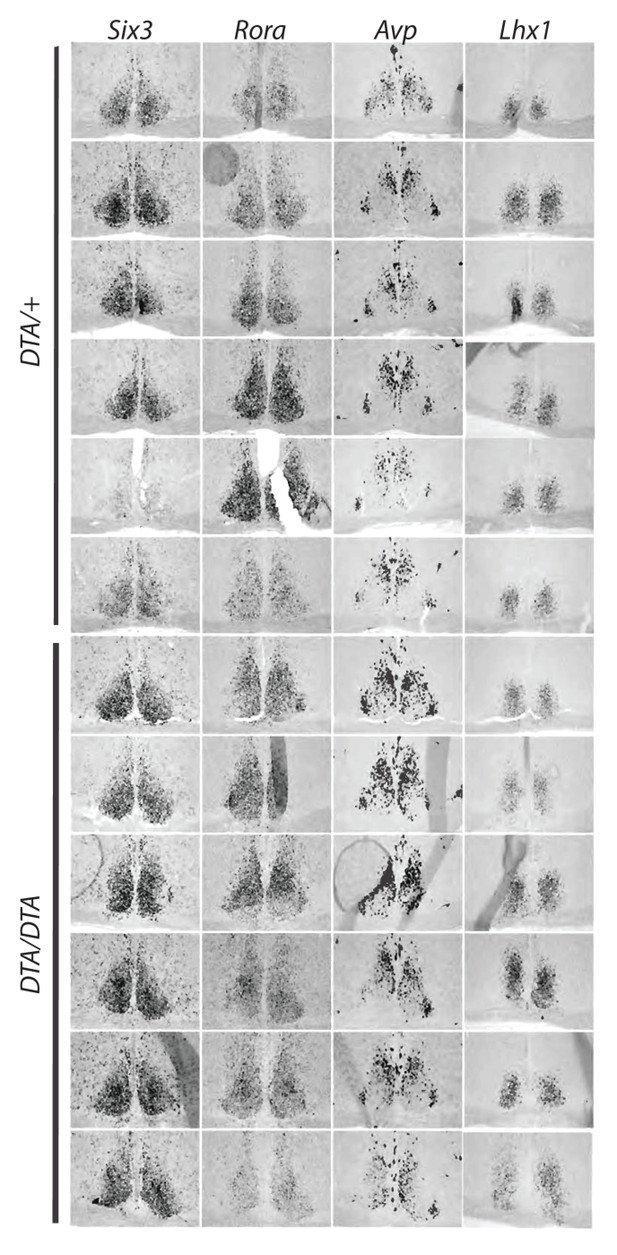
Expression pattern of transcription factors critical for SCN development.
In situ hybridization for several developmentally important transcription factors, Rora, Six3, and Lhx1 as well as the functionally important neuropeptide vasopressin (AVP) in control (Opn4DTA/+) and Opn4DTA/DTA mice to test whether the lengthened circadian period exhibited by Opn4DTA/DTA (but not Opn4DTA/+) mice could be attributed to gross changes in the SCN. Each row represents one animal. The expression patterns of these genes were similar between these two groups of mice.
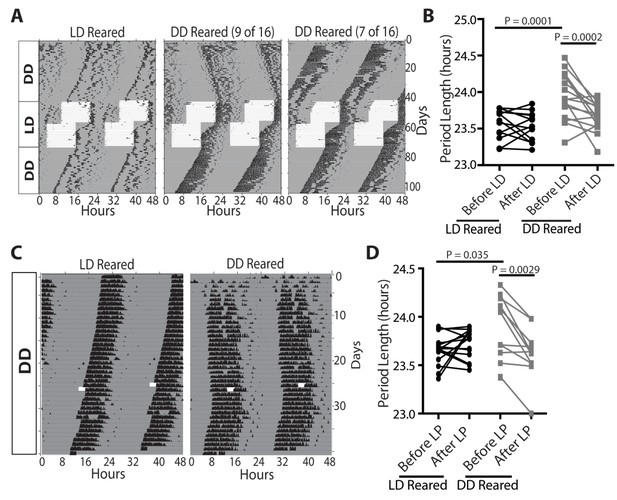
Light sets the circadian period length, even during adulthood.
(A and B) Representative actograms of wild-type mice raised in either a 12:12-LD cycle (n = 13) or darkness (n = 16), and exposed to 12:12-LD cycle for 1 month. The circadian period of dark-reared mice is lengthened initially, but rescued and is no longer significantly different from 12:12-LD reared mice after light exposure. (C and D) Representative actograms of wild-type mice raised in either a 12:12-LD cycle (n = 17) or in darkness (n = 12), and then exposed to a 3 hr light pulse. Dark-reared mice exhibited a longer period. Following the 3 hr light pulse, the circadian period length of dark-reared animals shortened. (B and D) Two-way ANOVA, Bonferroni's multiple comparisons test and adjusted p values. Error bars represent s.e.m. See also Figure 3—figure supplement 1.
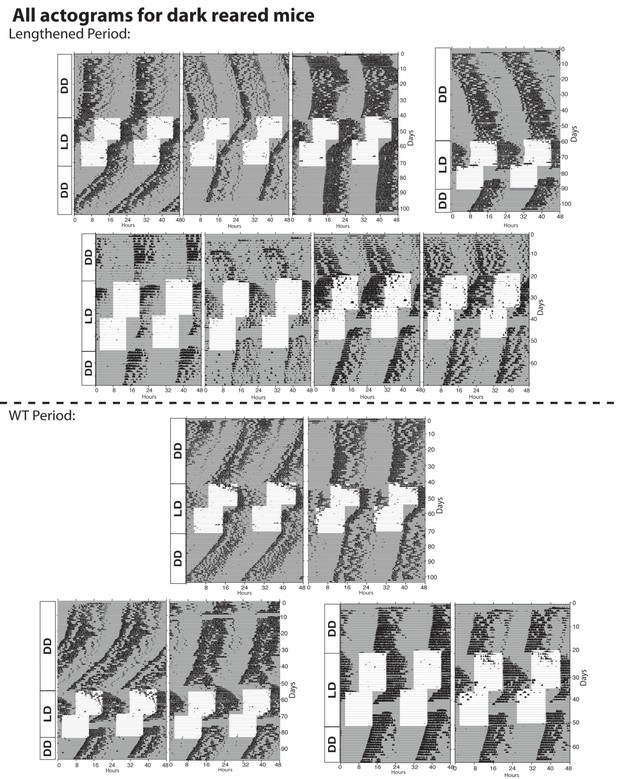
Actograms for all dark reared mice tested.
The lengthened period phenotype exhibited by dark-reared animals was less penetrant than in Opn4DTA/DTA or P0 enucleated mice. Though a majority of dark-reared animals (9 of 16; 56%;) exhibited a lengthened period, the remainder did not. This is likely due to the process of setting the circadian period being highly sensitive to light.
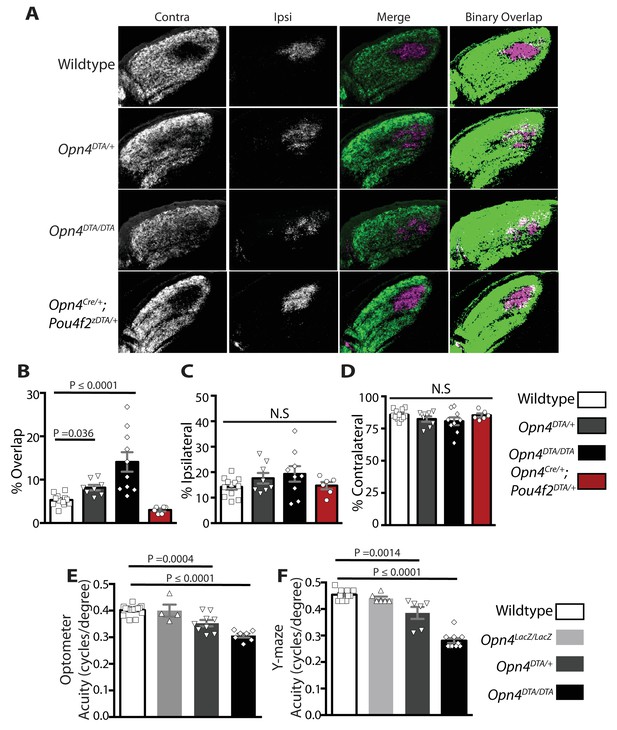
Adult Opn4DTA/DTA mice display deficits of eye-specific axonal segregation and visual acuity.
(A) RGC axonal innervation of the adult dLGN of wild-type (n = 12), Opn4DTA/+ (n = 8), Opn4DTA/DTA (n = 10) and Opn4Cre/+; Pou4f2zDTA/+ (n = 6; previously published as Opn4Cre/+; Brn3bzDTA/+). The rightmost images represent binarized version of the merged images to visualize the overlap between contralateral and ipsilateral RGC projections. Representative images were taken from the region of the dLGN indicated by the blue arrow in Figure 4—figure supplement 4F. (B) Opn4DTA/DTA mice exhibited a significantly higher percentage of overlapping pixels relative to the total number of LGN than any other tested genotype. Opn4DTA/+ mice exhibited levels of overlapping pixels that were intermediate compared to Opn4DTA/DTA and control mice. (C and D) The percentage of the total number of pixels in the dLGN from ipsilateral and contralateral fibers is similar among all tested genotypes. (E and F) The virtual optokinetic system and visual water task were used to assess visual function wild-type (virtual optokinetic system: n = 16, visual water task: n = 10), Opn4LacZ/LacZ (virtual optokinetic system: n = 4, visual water task: n = 6), Opn4DTA/+ (virtual optokinetic system: n = 9, visual water task: n = 6), Opn4DTA/DTA (virtual optokinetic system: n = 7, visual water task: n = 10) mice. Wild-type and Opn4LacZ/LacZ mice were indistinguishable. Opn4DTA/DTA mice exhibited reduced visual acuity compared to wild-type mice, and Opn4DTA/+ mice exhibited intermediate visual acuity. (B–F) One-way ANOVA, Bonferroni's multiple comparisons test and adjusted p values. Error bars represent s.e.m. See also Figure 4—figure supplements 1–5.
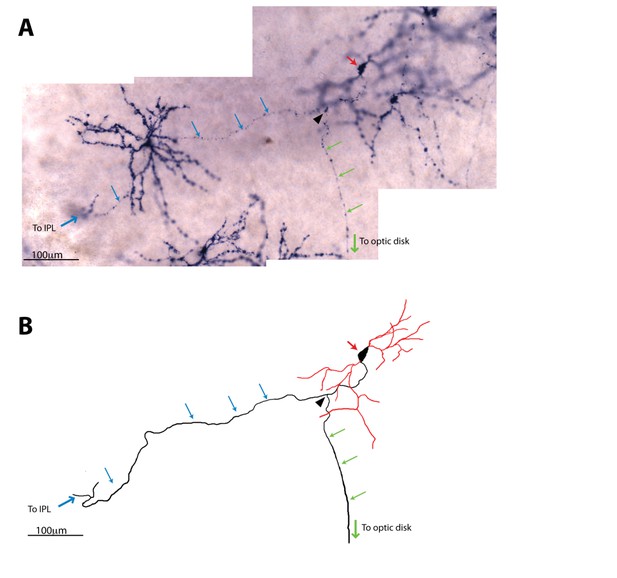
ipRGC intra-retinal axonal collaterals are present by P7.
(A) Alkaline phosphatase (AP) staining in P7 Opn4CreERT2/+ retinas (Chen et al., 2013; Joo et al., 2013) from mice that had been injected with a low dose of tamoxifen at P0, which resulted in sparse labeling of ipRGCs with AP. This image shows an ipRGC axon branching with one branch (green arrows) projecting to the optic disk and the other (blue arrows) to the inner plexiform layer (IPL). Red arrow indicates the cell body, and the black arrowhead indicates the branching point of the axon. (B) A trace of the ipRGC shown in (A) that projects an intra-retinal axon collateral. The trace was compiled from images of consecutive focal planes. Red lines indicate dendrites and black lines indicate the axon.
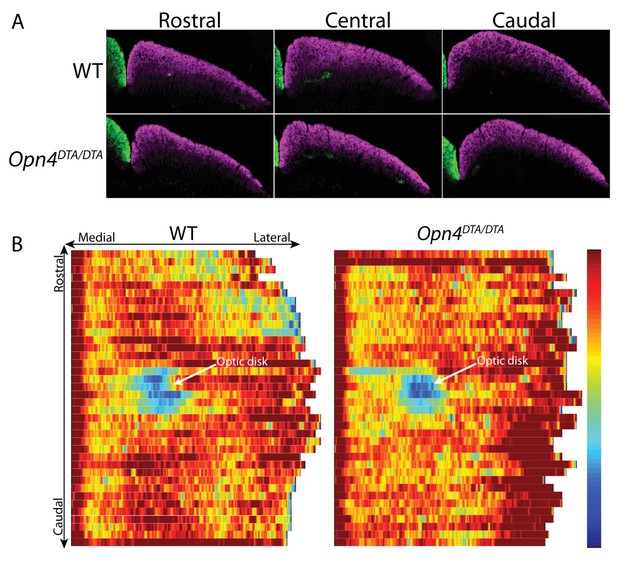
Retinal innervation of the SC in Opn4DTA/DTA mice is indistinguishable from wild-type mice.
(A) Representative images of RGC axonal innervation of the adult SC in WT and Opn4DTA/DTA mice. Alexa Fluor 594- (purple) and 488-(green) conjugated CTB were injected into the right and left eye, respectively, to label RGC projections. Images are of the left SC. (B) Heat map showing a reconstruction of the dorsal view of retinal innervation of the SC.
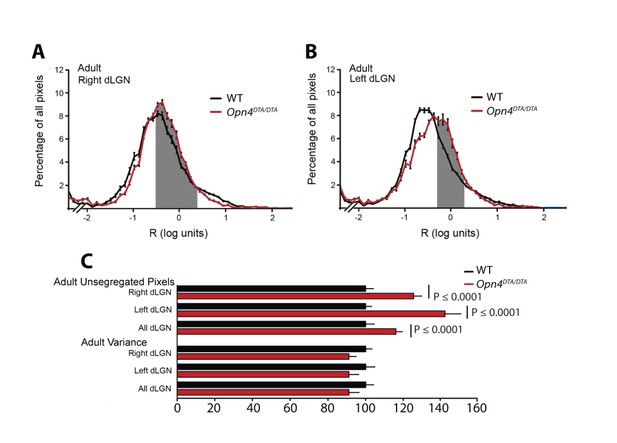
Quantification of disruption in eye-specific axonal segregation in the dLGN of adult Opn4DTA/DTA mice.
(A and B) The averaged distribution of pixel intensity ratios (R) for all dLGN pixels in the right (A) and left (B) dLGN of adult WT (black curve) and Opn4DTA/DTA (red curve) mice are plotted. The mice were raised in 12:12 LD cycle. Opn4DTA/DTA mice have more unsegregated pixels (gray shaded area-see methods) than WT mice. (C) Pooled data normalized to WT mice indicated Opn4DTA/DTA mice have 16% more unsegregated pixels, and a 9% decrease in the variance of the R distribution (n = 4 WT, 4 Opn4DTA/DTA, 15 sections/mouse/hemisphere). Student-t test. Error bars represent s.e.m.
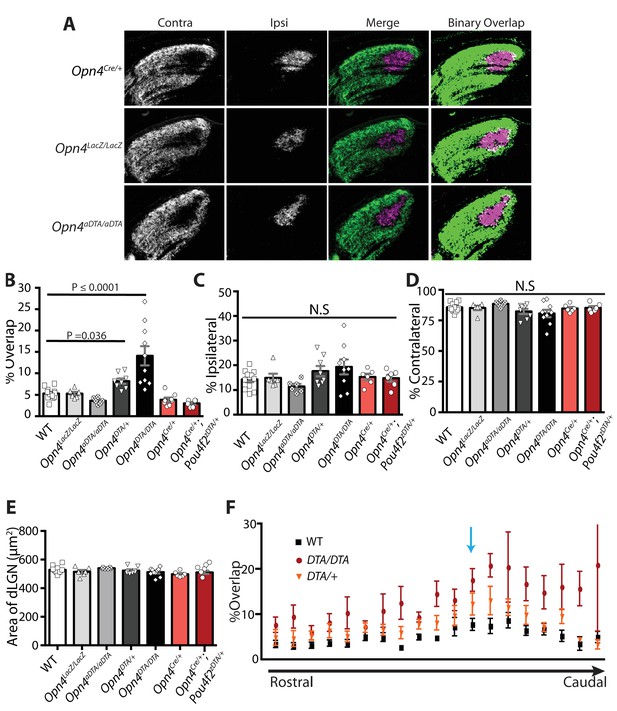
Eye-specific axonal segregation is normal in Opn4Cre/+, Opn4LacZ/LacZ, and Opn4aDTA/aDTA mice.
(A) RGC axonal innervation of adult dLGN in Opn4Cre/+, Opn4LacZ/LacZ, and Opn4aDTA/aDTA. Images are of the right dLGN. The rightmost images are a binarized version of the merged images to visualize the overlap between contralateral and ipsilateral RGC projections. Representative images were taken from the region of the dLGN indicated in D (blue arrow). Opn4LacZ/LacZ and Opn4aDTA/aDTA mice served as additional controls for Opn4DTA/DTA mice and were indistinguishable from WT. Opn4Cre/+ mice served as an additional control for Opn4Cre/+; Pou4f2zDTA/+ mice (previously published as Opn4Cre/+; Brn3bzDTA/+) and were indistinguishable from each other as well as not significantly different from WT mice. (B) Percentage of overlapping pixels in all tested genotypes. (C, D) The percentage of the total number of pixels in the dLGN from ipsilateral and contralateral fibers was similar among all tested genotypes. (E) Size of the dLGN is comparable across all tested genotypes. In B–D, values for WT, Opn4DTA/+, Opn4DTA/DTA and Opn4Cre/+; Pou4f2zDTA/+ mice were re-plotted from Figure 4B–D for comparison. * indicates p<0.05 and ** indicates p<0.001 with one-way ANOVA, Bonferroni's multiple comparisons test. (F) The rostral to caudal distribution of percent overlap between contralateral and ipsilateral projections through the dLGN. The blue arrow indicates the section of the dLGN presented in A) and in Figure 4A. Error bars represent s.e.m.
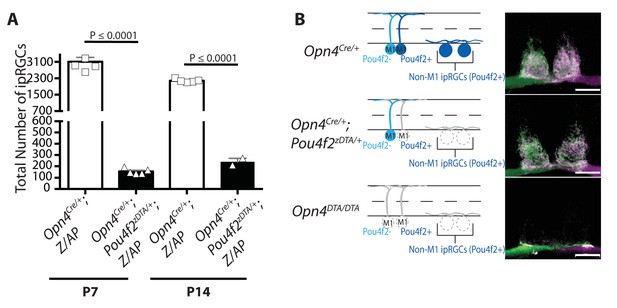
All but ~200 ipRGCs are ablated by P7 in Opn4Cre/+; Pou4f2zDTA/+ mice.
(A) Cell counts of total number of ipRGCs in Opn4Cre/+; Z/AP (control) and Opn4Cre/+; Pou4f2zDTA/+; Z/AP mice at P7 (control: n = 5; Opn4Cre/+; Pou4f2zDTA/+; Z/AP: n = 5) and P14 (control: n = 5; Opn4Cre/+; Pou4f2zDTA/+; Z/AP: n = 2). ipRGCs were identified by AP staining, which labels all subtypes. In Opn4Cre/+; Pou4f2zDTA/+; Z/AP mice, there was a significant loss of ipRGCs by P7 and no further loss at P14 suggesting that the loss of all but ~200 ipRGCs is complete by P7. One-way ANOVA Bonferroni's multiple comparisons test and adjusted p values. (B) Total retinal innervation of the SCN visualized with Alexa Fluor 594- (purple) and 488-(green) conjugated CTB in Opn4Cre/+ (control), Opn4Cre/+; Pou4f2zDTA/+, and Opn4DTA/DTA mice. Scale bar = 200 µm.
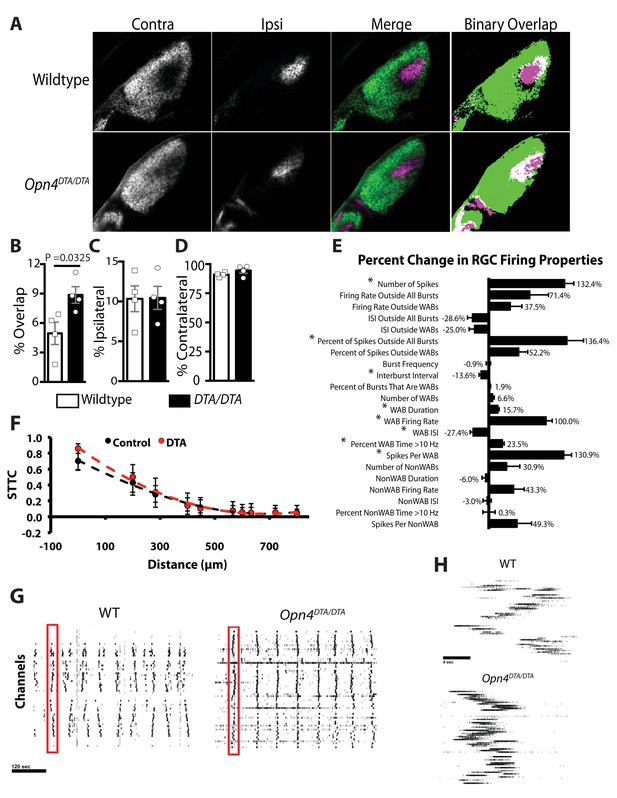
Opn4DTA/DTA mice display disrupted eye-specific axonal segregation as early as P8 and exhibit altered RGC firing properties.
(A) RGC axonal innervation of the dLGN in P8 wild-type (n = 4) and Opn4DTA/DTA (n = 4) mice as in Figure 3A. (B) At P8, Opn4DTA/DTA mice exhibit a significantly higher percentage of overlapping pixels relative to the total number of LGN pixels than wild-type mice. (C and D) The percentage of the total number of pixels in the dLGN from ipsilateral and contralateral fibers is similar among all tested genotypes. * indicates p<0.05 with Student’s t-test. Error bars represent s.e.m. for all graphs. (E) RGC spiking properties. Opn4DTA/DTA mice exhibit longer WABs and had a higher firing rate, with more spikes and a shorter inter-burst interval. There were also significantly more spikes outside of WABs. (F) Spike Time Tiling Coefficient (STTC) versus distance demonstrating that correlated spiking activity between neurons was similar in Opn4DTA/DTA mice compared to controls. 275 spike trains from wild-type/control, 245 from DTA. * represent statistically significant differences after Student's T-tests with a Holm-Bonferroni correction, α = 0.05, m = 22 for spiking properties, m = 10 for STTC.’ Exact p values are listed in Supplementary file 2. (G) Representative raster plots from multielectrode array recording of retinal waves in P6 WT and Opn4DTA/DTA mice in the dark. Each row represents the activity on a single electrode. (H) Expansion of one wave (identified by the red box in (G)). Error bars represent s.e.m. for all graphs.
Additional files
-
Supplementary file 1
Description of all mouse lines.
- https://doi.org/10.7554/eLife.22861.017
-
Supplementary file 2
Values and statistics for retinal wave recordings.
Properties of spontaneous retinal activity in P6 WT and Opn4DTA/DTA mice. Recordings were done in darkness. * represent statistically significant differences after Student's T-tests with a Holm-Bonferroni correction, α = 0.05, m = 22. Significance is determined when the p-value is less than or equal to the Holm-Bonferroni corrected p-value.
- https://doi.org/10.7554/eLife.22861.018





