Action of CMG with strand-specific DNA blocks supports an internal unwinding mode for the eukaryotic replicative helicase
Figures
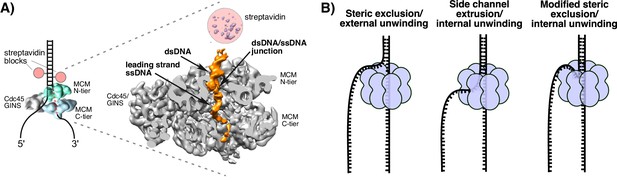
Structure of S. cerevisiae CMG at a replication fork and models of hexameric helicase unwinding.
(A) CryoEM single particle 3D reconstruction of active CMG that was captured by streptavidin blocks (left diagram); the CMG is a surface rendering. The right panel is the CMG-forked DNA complex as a cut-open surface rendering. Adapted from Figure 6 of Georgescu et al. (2017). (B) Models of hexameric helicase function. Left: classic steric exclusion in which the helicase encircles only one strand, excluding the other and the unwinding point is external to the helicase. Middle: classic side channel extrusion model with duplex DNA entering the channel and the DNA split point is internal, with one strand extruded out a side channel. Right: Proposed modified steric exclusion model with duplex entering the channel and an internal unwinding point, followed by exclusion of the non-tracking strand.
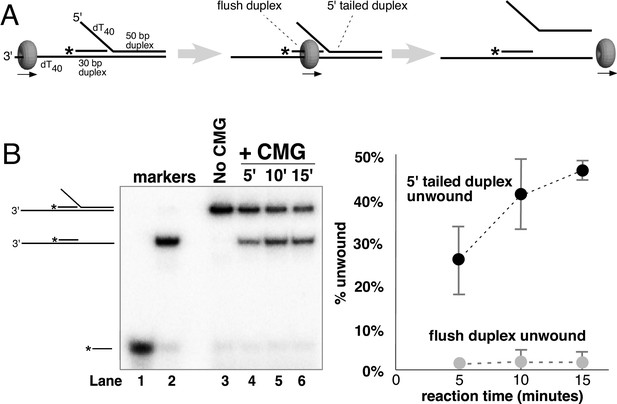
CMG translocates over flush duplex DNA without unwinding.
(A) The substrate contains a 3’ dT40 ssDNA tail for CMG loading and a 5’-32P labeled (*) flush duplex adjacent to a 5’ dT40 tailed duplex. CMG (grey ring) tracks 3’−5’ along ssDNA as indicated by the arrow in the schematic, unwinding the tailed duplex (right) but leaving the flush duplex in place. A detailed description of the substrate is shown in Figure 2—figure supplement 1; oligo sequences are in Table 1. (B) Left: native PAGE analysis of the CMG unwinding reaction. See Materials and methods for reaction conditions and details. Markers (lanes 1–3) show the positions of the species indicated to the left. Right: The plot shows the time course of unwinding of the tailed duplex (dark circles) and flush duplex (light circles). Values are the average of three independent experiments and the error bars show the standard deviation. Also see Figure 2—figure supplement 2.
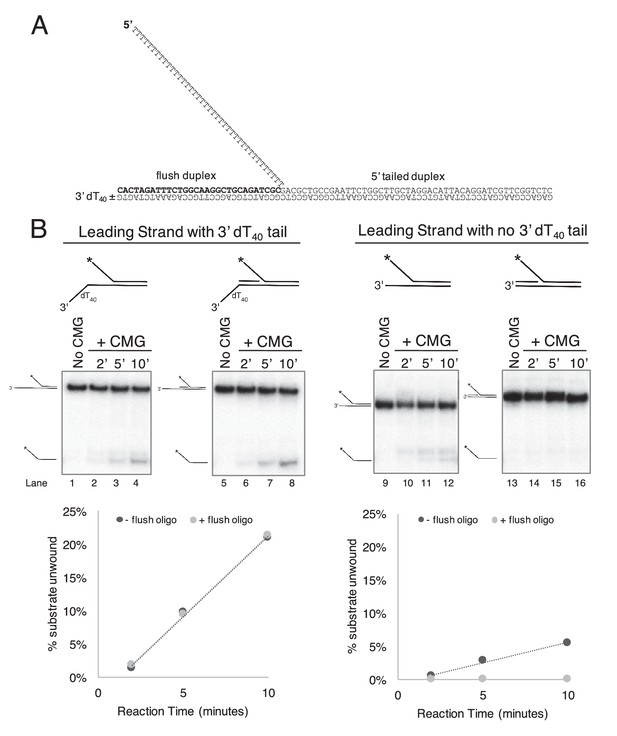
CMG requires a 3’ dT40/ssDNA tail for loading.
. (A) Schematic of the substrates used in Figure 2 and in part (B) of this Figure. Further details on the oligos used are in Table 1. The flush duplex oligo is named ‘flush duplex LAG’. The 5’ tailed oligo is ‘50duplex LAG’. The leading strand template is either ‘Paired duplex LEAD + 3’ tail’ or ‘Paired duplex LEAD no 3’ tail’. (B) Control experiments to show the requirement for the 3’ dT40 tail in CMG loading. At left are two helicase assays like those described in Figure 2 but using the substrate containing a 3’ dT40 tail with the radiolabel on the 5’ tailed duplex. Lanes 1–4 show unwinding in the absence of the flush duplex oligo and lanes 5–8 in the presence of the flush duplex. The % of 5’ tailed oligo unwound is quantified in the graph below the gels, showing that the presence of the flush duplex oligo does not affect CMG loading or unwinding. At right is an identical pair of experiments but with a substrate that does not contain the 3’ dT40 tail. Lanes 9–12 and the quantitation in the graph below show that unwinding is greatly reduced in the absence of the 3’ tail (compare lanes 9–12 with lanes 1–4), and in the presence of the flush duplex oligo (lanes 13–16) CMG does not load/unwind at all. These experiments support the conclusion of Figure 2 that CMG translocates over flush duplex without unwinding and requires a 3’ dT40 tail for efficient loading.
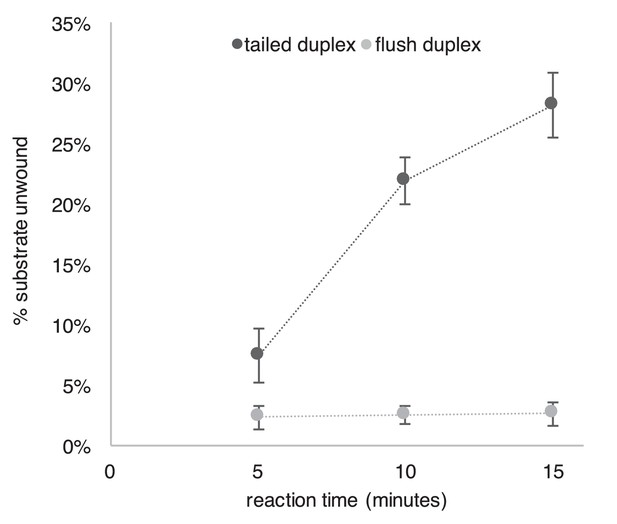
Repeat of Experiment from Figure 2B in the presence of a trap to prevent reannealing of flush duplex oligo.
The experiment from Figure 2B was repeated exactly as described in the Figure Legend except that 2 min after the reaction was started, a 40-fold excess of unlabeled flush duplex oligo was added (to a final concentration of 20 nM) to prevent reannealing of any unwound radiolabeled flush duplex oligo (0.5 nM). This experiment supports the conclusion of Figure 2 that CMG translocates over flush duplex without unwinding.
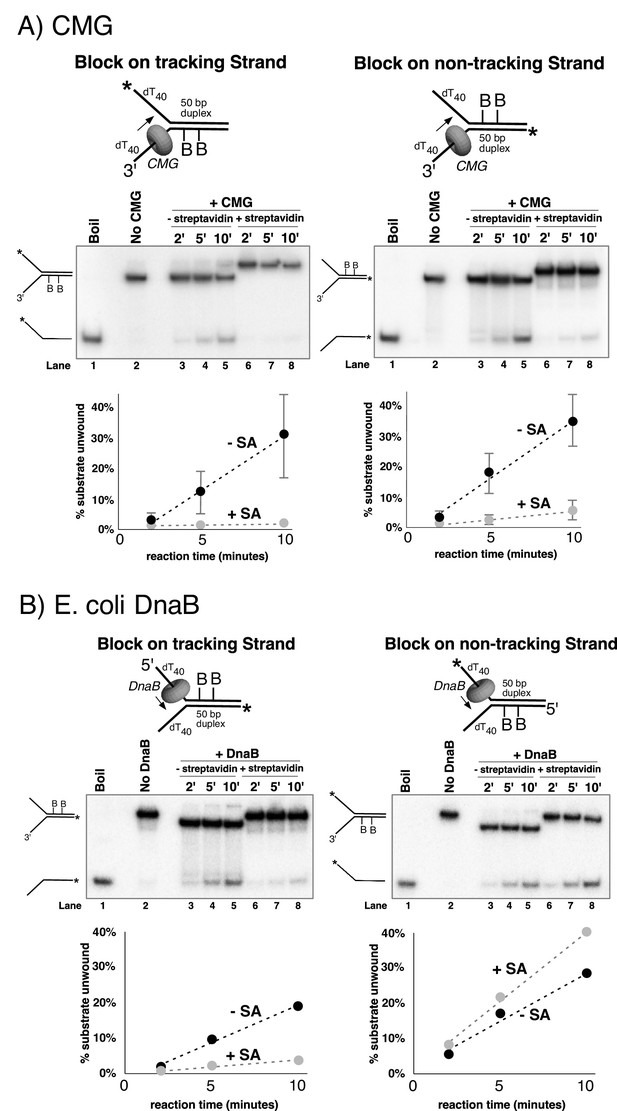
Dual biotin-streptavidin on either strand is a strong block to CMG unwinding.
(A) Effect of dual biotin blocks on CMG. CMG was mixed with dual biotinylated DNA fork and ATP in the absence (lanes 3–5) or presence (lanes 6–8) of streptavidin. The reaction is described in Materials and methods, and the substrates are shown in Figure 3—figure supplement 1 and in schematic above the gels. CMG (ring in the schematic) tracks 3’−5’ as indicated by the arrow. The radiolabeled strand is indicated by a * at its 5’ end. Lane 1 shows the position of the unwound radiolabeled strand (by boiling) and lane 2 shows the forked DNA. The plots below the gels show % substrate unwound at the 2’, 5’ and 10’ time points in the absence (dark circles) and presence (light circles) of streptavidin. Values are the average of three independent experiments and the error bars show the standard deviation. The dotted line is a linear least squares fit of the data. Left: the leading strand template contains dual biotin. Right: the lagging strand template contains dual biotin. Also see Figure 3—figure supplements 1–6. (B) Effect of dual biotin blocks on E. coli DnaB. As a control, we used E. coli DnaB, known to act by classic steric exclusion/external unwinding (Kaplan, 2000; Kaplan et al., 2003). DnaB translocates 5’−3’ on ssDNA, placing it on the lagging strand. Left: dual biotin on the lagging strand is a strong block to DnaB unwinding when streptavidin is present (lanes 6–8) compared to the no streptavidin control (lanes 3–5). Right: dual biotin on the leading strand does not inhibit DnaB in the presence (lanes 6–8) or absence of streptavidin (lanes 3–5).
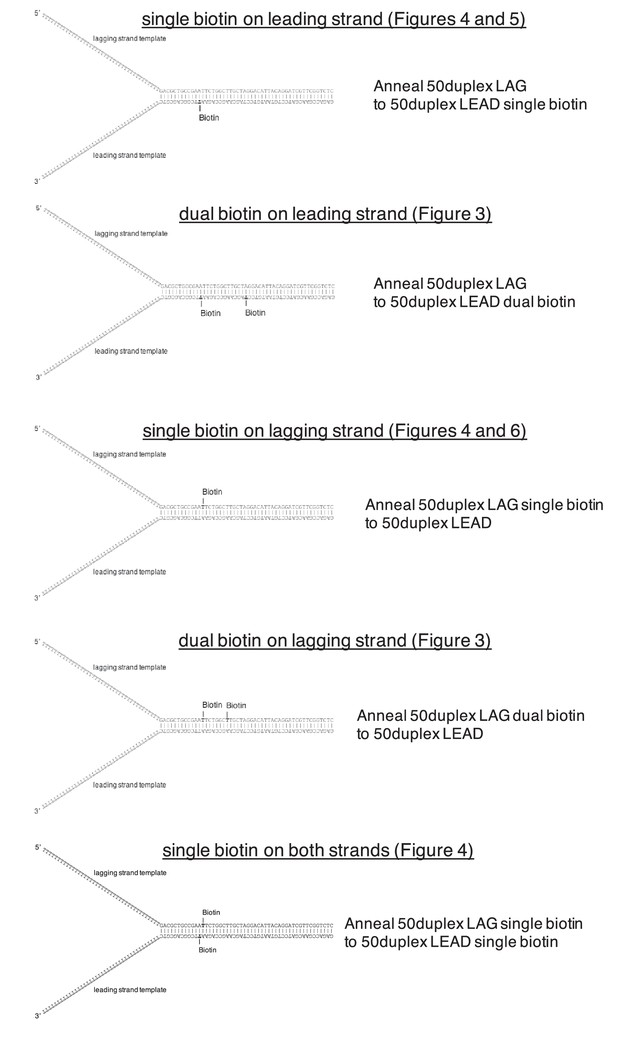
Schematics of biotinylated DNA fork substrates.
The fork substrates used in the experiments in Figures 3–6 are shown including the location of the biotinylated dT nucleotides in either the leading or lagging strand template. The oligos used to make the forked duplex substrates are indicated to the right of the schematics and the oligo sequences and modifications are in Table 1.
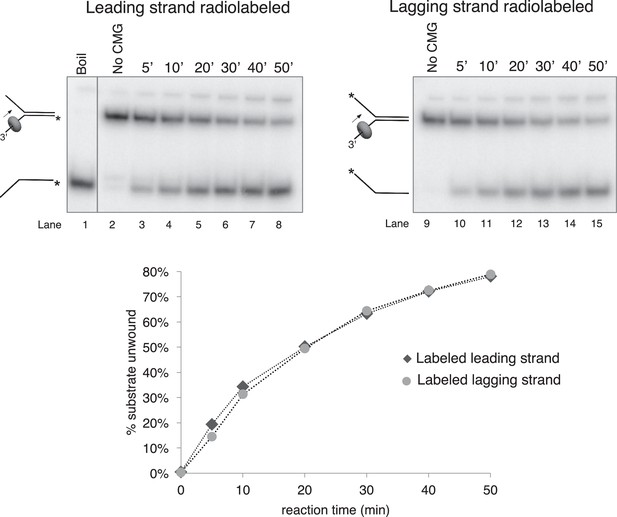
Time course of CMG unwinding on forked duplex.
Time course of unwinding of the forked duplex substrate like those shown in Figure 3—figure supplement 1 but with no biotin modifications. Lane 1 shows the position at which the unwound radiolabeled strand migrates (by boiling of the duplex substrate), lane 2 shows the migration of the substrate in the absence of CMG, and lanes 3–8 show substrate unwinding by CMG over a 50’ time course. The % of the radiolabeled strand unwound is shown in the graph at bottom. Lanes 9–15 are a repeat of this experiment with the radiolabel on the opposite strand, showing that unwinding is not affected by the position of the radiolabel. Reactions were mixed on ice and contained 0.5 nM radiolabeled DNA substrate and 20 nM CMG in a total reaction volume of 90 μl. Reactions were started by incubating at 30°C and 12 μl aliquots were removed, stopped with EDTA/SDS, and flash frozen in liquid nitrogen at the time points indicated in the Figures to ensure no reannealing takes place. Reaction products were separated on 10% Native PAGE minigels and subsequently exposed to a storage phosphor screen that was scanned on a Typhoon 9400 laser imager (GE Healthcare). Scanned gels were analyzed using ImageQuant TL v2005 software.
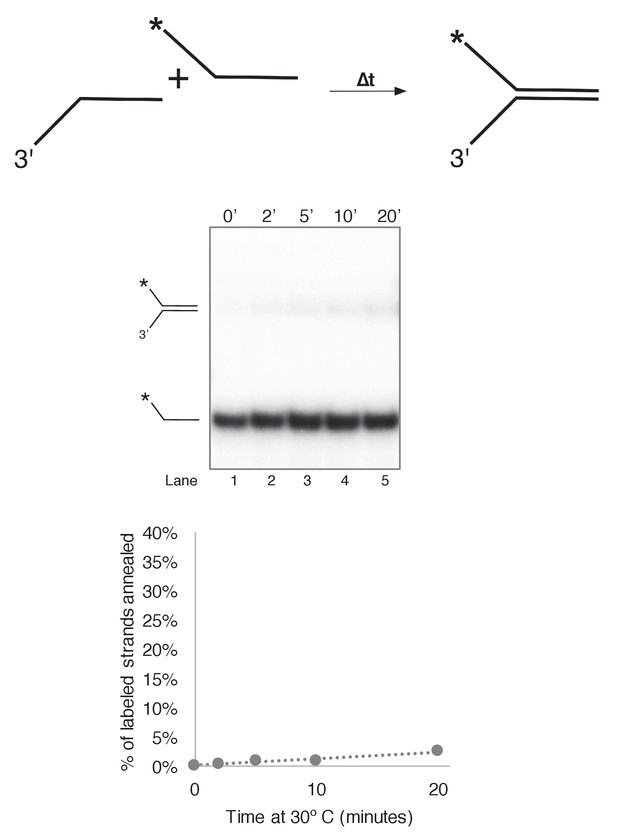
Substrate single strands do not spontaneously reanneal at 30˚C.
To determine whether unwound substrate oligos spontaneously reanneal under our reaction conditions, we mixed 0.5 nM final concentration of radiolabeled 50duplex LAG oligo (Table 1) with 0.5 nM unlabeled 50 duplex LEAD oligo under reaction conditions identical to those of Figure 3 in a total reaction volume of 55 μl. Complete reactions were mixed on ice and started by incubating at 30°C. At the indicated time points, 10 μl aliquots were removed and flash frozen in liquid nitrogen. Lack of re-annealing of the unwound strands allowed us to perform unwinding reactions in Figure 3 and subsequent Figures in the absence of a trap, eliminating potential side interactions between CMG and the unlabeled trap DNA.
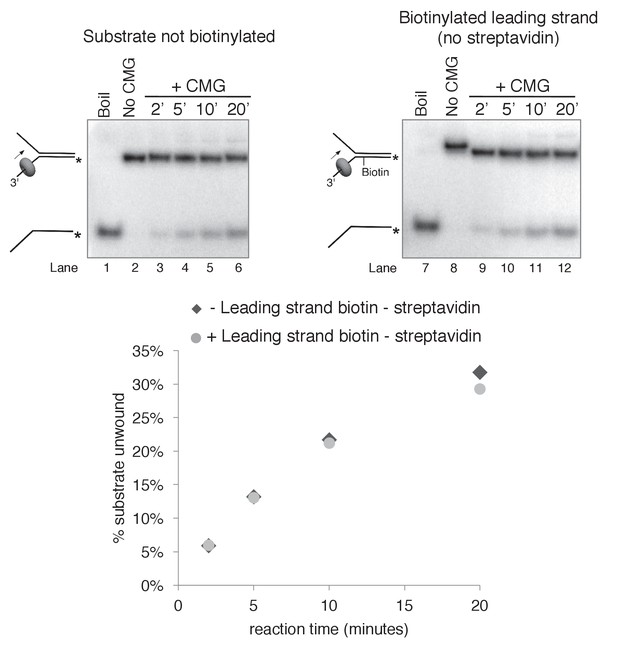
Biotinylation of the substrate does not affect CMG unwinding in the absence of streptavidin.
As a control to determine whether the biotin modification on the substrate affects CMG unwinding in the absence of streptavidin, we compared CMG unwinding of a non-biotinylated substrate (lanes 3–6) to that of the substrate with biotin on the leading strand template (lanes 9–12). Reactions conditions were identical to those in Figure 3—figure supplement 2 except the total reaction volume was 60 μl and 12 μl time points were taken over a time course out to 20’. As shown in the graph at bottom, the amount of substrate unwound was identical in the absence and presence of the biotin modification.
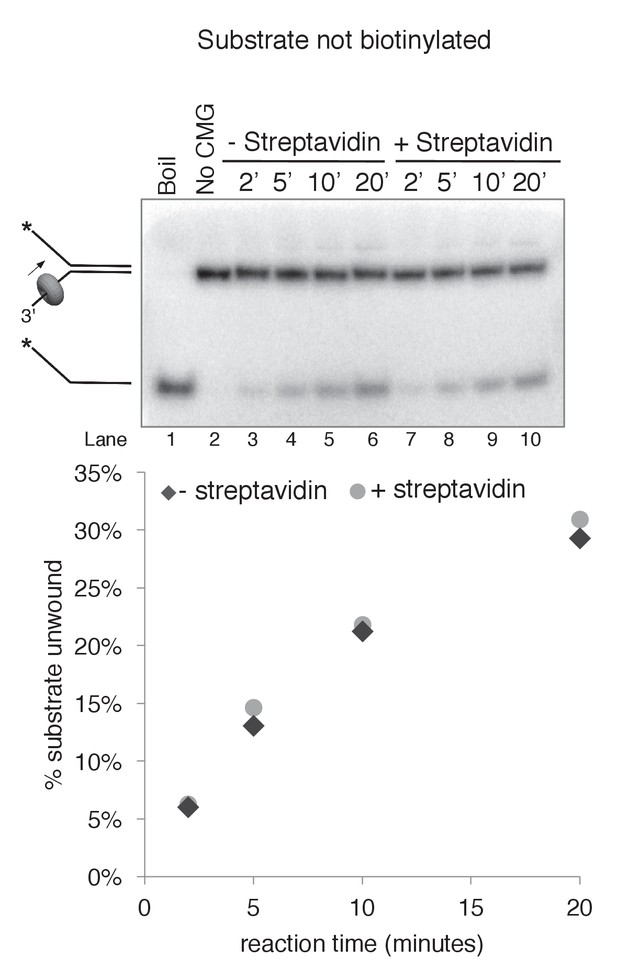
Streptavidin does not affect CMG unwinding of a non-biotinylated substrate.
As a control to determine whether addition of streptavidin affects CMG unwinding of a non-biotinylated substrate (i.e. non-specific inhibition of unwinding), we compared CMG unwinding of a non-biotinylated substrate in the absence (lanes 3–6) and presence (lanes 7–10) of streptavidin. Reactions conditions were identical to those in Figure 3—figure supplement 2 except for the addition of 2 μg/ml (final concentration) streptavidin to the reaction in lanes 7–10; the total reaction volume was 60 μl and 12 μl time points were taken over a time course out to 20’. As shown in the graph at bottom, the amount of substrate unwound was identical in the absence and presence of streptavidin when the substrate was not biotinylated.
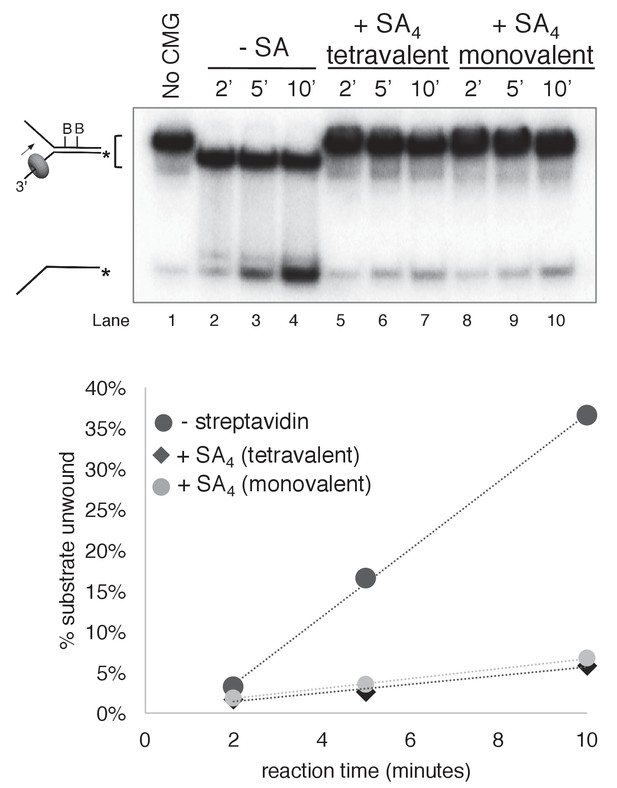
Monovalent streptavidin (SA) tetramer inhibits CMG unwinding to the same extent as tetravalent streptavidin on the substrate with dual biotin on the lagging strand.
Three helicase assays were performed under otherwise identical conditions comparing CMG unwinding in the absence of streptavidin (lanes 2–4); in the presence of the tetravalent streptavidin used in the experiments of Figures 3–6 (lanes 5–7); and in the presence of a monovalent form of streptavidin in which only one of the four protomers of the streptavidin tetramer binds to biotin while retaining the high affinity of the tetravalent form (Howarth et al., 2006). As shown in the graph below the gel, monovalent streptavidin inhibits CMG unwinding to the same extent as the standard tetravalent form, indicating that inhibition of the helicase is not attributable to distortion of the duplex from ‘cross-linking’ of two nearby biotins by a single streptavidin tetramer. In these experiments, each reaction contained 20 nM CMG in a total reaction volume of 45 μl with 12 μl samples stopped with EDTA/SDS at the times indicated in the Figure. For the reaction in lanes 5–7, 4 μg/ml (final concentration) tetravalent streptavidin was incubated with the substrate at 30°C for 5’ and then on ice for 10’ before addition of CMG. For the reactions in lanes 8–10, 0.5 μg/ml (final concentration) of monovalent streptavidin was used instead.
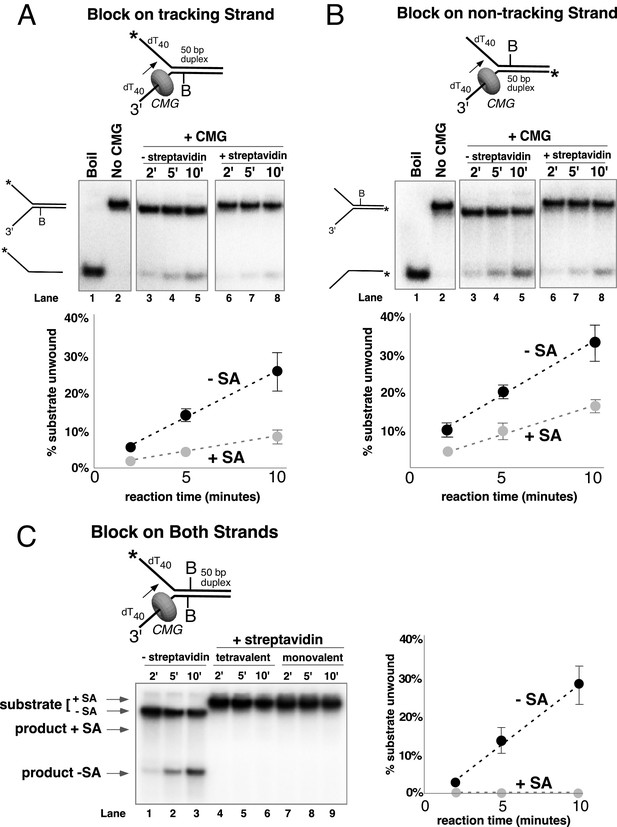
Single biotin-streptavidin is a weaker block to CMG unwinding on either strand but not when placed on both strands.
Reaction conditions and analysis are identical to those in Figure 3 except that the substrate contained a single biotin on the leading strand (A), lagging strand (B), or both (C) as indicated in the schematics above the gels; and the substrate in (A) and (B) was pre-incubated with 2 μg/ml streptavidin before addition of CMG instead of 4 μg/ml. The substrates are described in detail in Figure 3—figure supplement 1.
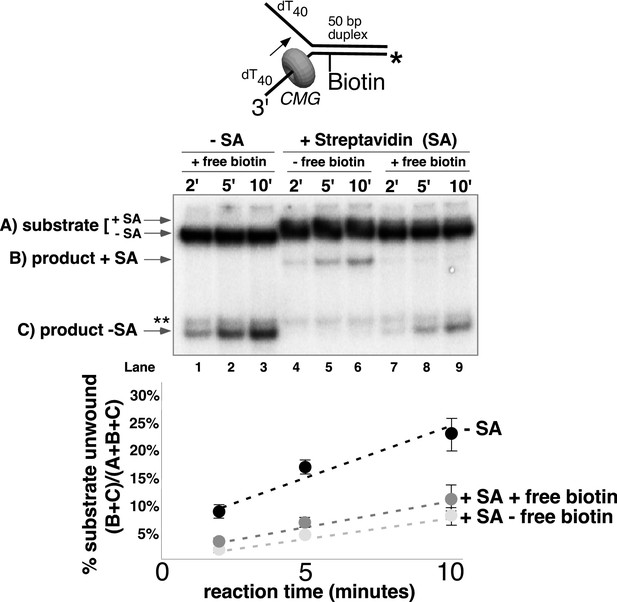
CMG can displace streptavidin from biotin on the leading strand.
In the reaction shown in lanes 4–9, the substrate was pre-incubated with 2 μg/ml streptavidin for 5’ at 30°C and then placed on ice for 10’ before addition of CMG only (lanes 4–6) or CMG plus D-biotin to a final concentration of 750 nM (lanes 7–9) as a trap for streptavidin displaced from biotin by CMG. The reaction in lanes 1–3 was performed in the absence of streptavidin. As shown in the schematic above the gel, in these experiments the radiolabel (*) was on the leading strand template containing the biotin-dTTP and when unwound it migrates at different positions in the gel in the presence (B) or absence (C) of streptavidin (SA) as indicated to the left of the gel. The band at the position indicated by ** to the left of the gel is a background band that is observed when the biotinylated strand is radiolabeled. Below the gel is a plot of the time course of unwinding for: lanes 1–3 (-SA), lanes 4–6 (+SA – free biotin), and lanes 7–9 (+SA + free biotin) showing the percent unwound product relative to total DNA (product plus unreacted substrate). Values are the average of three independent experiments and the error bars show the standard deviation. Displacement of streptavidin by CMG is revealed by a down shift in the migration of the unwound product from the product +SA position (B) in the absence of the D-biotin trap (lanes 4–6) to the product –SA position (C) in the presence of the trap (lanes 7–9). Also see Figure 5—figure supplement 1.
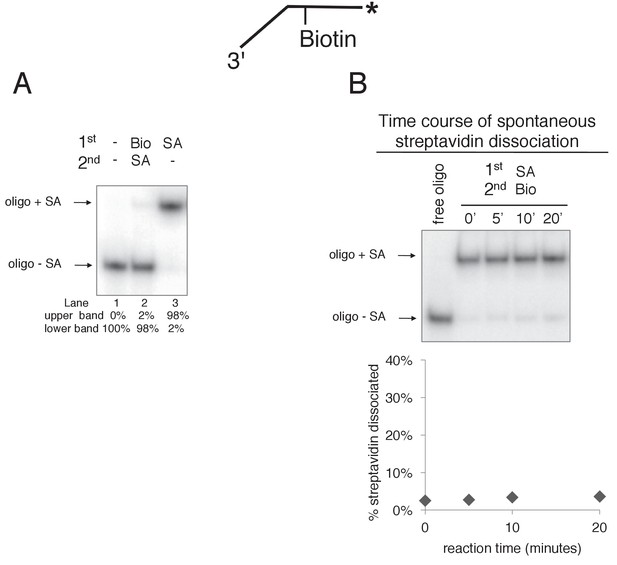
Free biotin trap prevents binding of streptavidin to biotinylated oligo when added before streptavidin but not when added after streptavidin.
(A) To validate the use of free biotin as a trap for displaced streptavidin, we examined the migration of the biotinylated leading strand ssDNA (50duplex LEAD single biotin, see Table 1) in the absence of streptavidin (lane 1), when biotin is added before streptavidin (lane 2), and when streptavidin is added without biotin (lane 3). The radiolabeled substrate was present at 0.5 nM and streptavidin and free biotin were at 2 μg/ml and 750 nM, respectively (final concentrations). The substrate was pre-incubated at 30°C for 5’ with biotin (lane 2), streptavidin (lane 3) or neither (lane 1) and then placed on ice for 10’. Then streptavidin was added to the reaction in lane 2 and all three reactions were incubated at 30°C for 10’ before loading on the gel. (B) The biotin trap does not displace streptavidin from biotin. As in (A), the substrate was pre-incubated with streptavidin before addition of free biotin and then incubated at 30°C. Samples were withdrawn from the reaction at the indicated times, quenched with SDS/EDTA and flash frozen in liquid nitrogen. The graph below the gel shows that dissociation of streptavidin is negligible over a 20’ time course in the presence of the biotin trap.
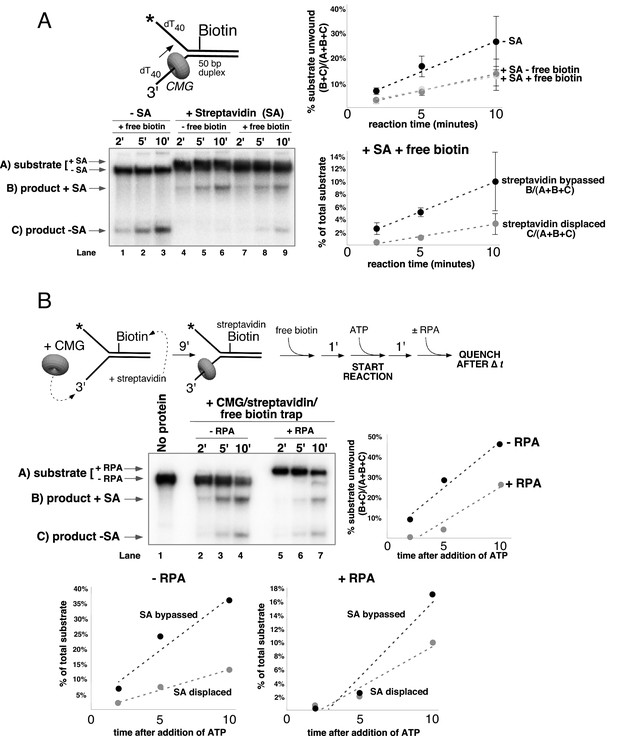
CMG can bypass or displace streptavidin from the lagging strand.
(A) Reaction conditions and analysis are identical to those in Figure 5 except the substrate was radiolabeled (*) on the biotinylated lagging strand template as shown in the schematic above the gel. Reactions in the absence of biotin trap (lanes 4–6) show the upshifted unwound product DNA. Reactions in the presence of biotin trap (lanes 7–9) show two products. One product is in the upshifted position and thus still contains streptavidin, and the other product is downshifted to the position of unwound substrate lacking bound streptavidin (SA displaced). The plot at the top right shows total unwinding in the absence of streptavidin (lanes 1–3) and in the presence of streptavidin with free biotin (lanes 7–9) or without (lanes 4–6). Values are the average of three independent experiments and the error bars show the standard deviation. The plot at the bottom right shows the time course of appearance of the distinct SA bypassed and SA displaced products in lanes 7–9. Also see Figure 6—figure supplements 1–2. (B) Single hit experiment using RPA to prevent reinitiation of CMG loading during the assay. Top: scheme of the experiment. CMG is preincubated 9 min with the forked DNA having a single lagging strand biotin-streptavidin block, then biotin trap is added, followed by ATP, and RPA is added 1 min later. The gel, below, shows equivalent reactions except RPA was not added to the reaction in lanes 2–4. Comparison of reactions without RPA (lanes 2–4) with reactions containing RPA (lanes 5–7) shows that RPA blocks reinitiation, as demonstrated previously (Georgescu et al., 2014). The plots below the gel show the quantitation of the two products formed in reactions minus RPA (left) and plus RPA (right).
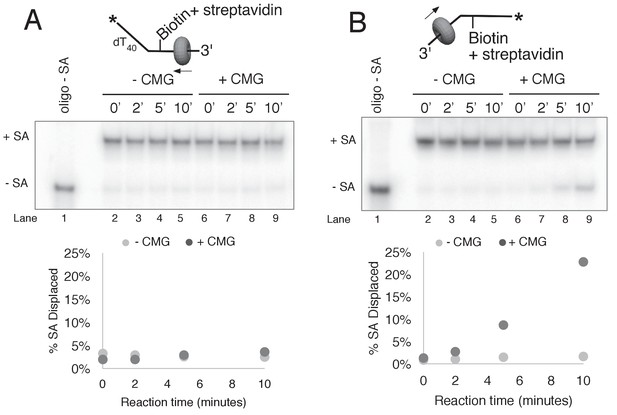
CMG displacement of streptavidin from ssDNA.
The ability of CMG to remove streptavidin from the lagging strand ssDNA (A) and leading strand ssDNA (B) was examined. In order to remove streptavidin from an ssDNA oligo, CMG must load onto the 3’ end and translocate along ssDNA in order to displace streptavidin from biotin, presumably by the force of translocation. As such, this assay examines the efficiency with which CMG loads onto the 3’ end of a given ssDNA. The radiolabeled ssDNA substrates (0.5 nM) were pre-incubated at 30°C for 5’ in the presence of 2 μg/ml streptavidin and then placed on ice for 10’ before addition of 25 nM CMG and 750 nM free biotin (final concentrations) to a total reaction volume of 60 μl. Reactions were started by incubating at 30°C and 12 μl aliquots were removed, stopped with EDTA/SDS, and flash frozen in liquid nitrogen at the time points indicated in the Figures. (A) The ability of CMG to remove streptavidin from lagging strand template ssDNA was examined. The substrate is 50duplex LAG single biotin (see Table 1) radiolabeled at the 5’ end. Lane 1 shows the migration of the substrate in the absence of streptavidin and lanes 2–5 show a time course of spontaneous dissociation of streptavidin in the presence of the trap (no CMG). Lanes 6–10 show removal of streptavidin by CMG translocating along the ssDNA substrate in the presence of the trap. As shown in the graph below the gel, both spontaneous dissociation (- CMG) and active removal of streptavidin (+ CMG) are negligible over the 10’ time course. (B) The ability of CMG to remove streptavidin from leading strand ssDNA was examined. The reactions in (A) were repeated with 50duplex LEAD single biotin (Table 1) radiolabeled at the 5’ end as substrate. As shown in the graph below the gel, CMG removes streptavidin much more efficiently from leading strand ssDNA than from lagging strand ssDNA (A). Based on the results of Figure 2—figure supplement 1, we presume that the presence of dT40 at the 3’ end of the leading strand oligo confers better loading than the dN50 at the 3’ end of the lagging strand oligo in (A).
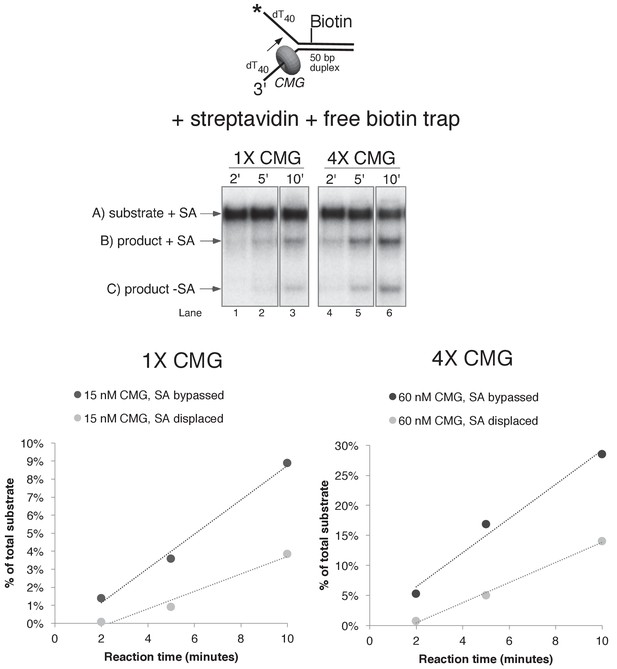
Helicase reactions by CMG on a single lagging strand biotinylated fork using two concentrations of CMG.
The reaction from lanes 7–9 in Figure 6A (with streptavidin and the free biotin trap) was repeated at two different concentrations of CMG and the results are shown in the gels at the top. Below the gels are plots of the two individual products that form, product with streptavidin bound, and products without streptavidin bound (streptavidin displaced).
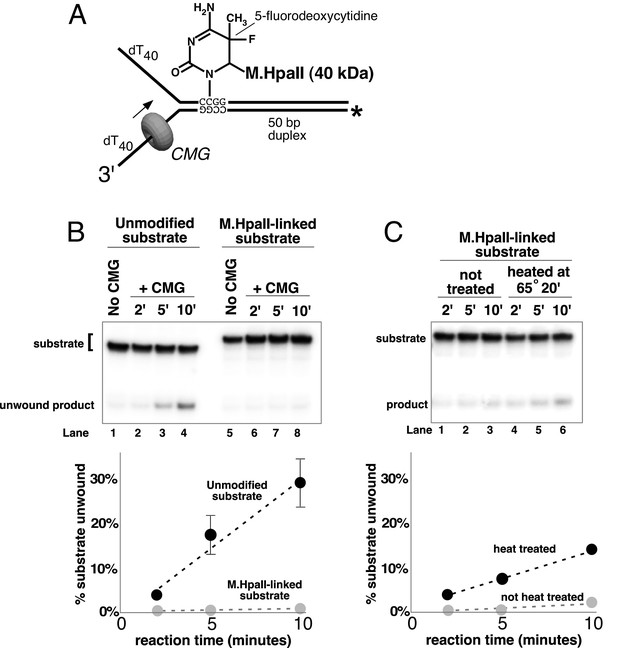
A covalent lagging strand protein-DNA adduct forms a stringent block to CMG.
(A) Schematic of the substrate used in these reactions. The duplex portion of the fork contains the 4-base recognition sequence for the HpaII methyltransferase. Replacement of the second dCTP in the recognition site with 5-fluorodeoxycytidine (5-FDC) traps a covalent intermediate in the methylation reaction in which M.HpaII remains bound to the nucleotide base on the lagging strand as shown (Chen et al., 1991). Sequences of the oligos are in Table 1. (B) CMG unwinding reactions using the 5-FDC substrate with (lanes 6–8) or without (lanes 2–4) M.HpaII modification. Except for the substrate, reactions conditions are the same as those of Figure 3 (no streptavidin). (C) The M.HpaII modified 5-FDC substrate was heated at 65° for 20’ to inactivate M.HpaII, cooled on ice, and added to a CMG unwinding reaction (lanes 4–6) identical to that in (B) except 40 nM CMG was used. Lanes 1–3 show unwinding of the untreated substrate under the same conditions.
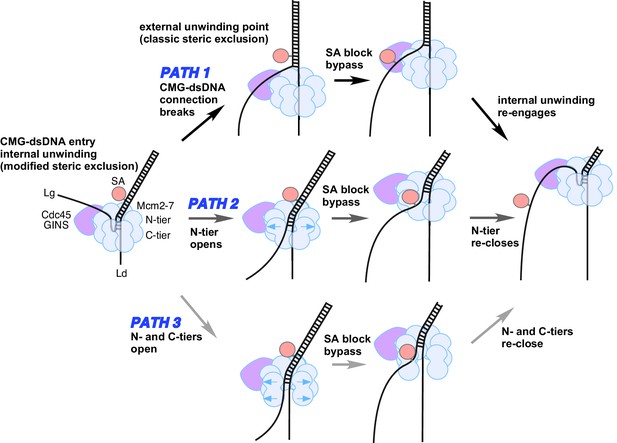
Possible paths of CMG bypassing streptavidin blocks on the lagging strand.
The illustration at the far left depicts the observed structure of CMG at a fork and the proposed exit path of the lagging strand template. Path 1 illustrates conversion to a classic steric exclusion process with an external unwinding point. Paths 2 and 3 illustrate streptavidin bypass by opening of either the N-tier or both tiers of the Mcm2-7 ring. The unwinding point could remain internal, as illustrated, or could become external. The illustration at the right suggests the CMG reassumes its initial conformation after passing the block.
Tables
Oligonucleotides used in this study. All oligonucleotides used in this study were ordered from IDT with the indicated modifications.
| Oligo name | Sequence (5’ to 3’) | Modification(s) |
|---|---|---|
| Paired duplex LEAD + 3’ tail | GAGACCGAACGATCCTGTAATGTCCTAG CAAGCCAGAATTCGGCAGCGTCGCGATC TGCAGCCTTGCCAGAAATCTAGTGTTTT TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT | - |
| Paired duplex LEAD no 3’ tail | GAGACCGAACGATCCTGTAATGTCCTAG CAAGCCAGAATTCGGCAGCGTCGCGATC TGCAGCCTTGCCAGAAATCTAGTG | - |
| flush duplex LAG | CACTAGATTTCTGGCAAGGCTGCAGATCGC | - |
| 50duplex LAG | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT TTTTTTTTTGACGCTGCCGAATTCTGGCTT GCTAGGACATTACAGGATCGTTCGGTCTC | - |
| 50duplex LAG single biotin | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT TTTTTTTTTGACGCTGCCGAATTCTGGCTT GCTAGGACATTACAGGATCGTTCGGTCTC | Single biotin-modified dT nucleotide in BOLD |
| 50duplex LAG dual biotin | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTT TTTTTTTTTTGACGCTGCCGAATTCTGGCTTG CTAGGACATTACAGGATCGTTCGGTCTC | Two biotin-modified dT nucleotides in BOLD |
| 50duplex LEAD | GAGACCGAACGATCCTGTAATGTCCTAG CAAGCCAGAATTCGGCAGCGTCTTTTTT TTTTTTTTTTTTTTTTTTTTTTTTTTTT*T*T*T*T*T*T | The six dT nucleotides at the 3’ end are connected by phosphorothioate bonds (*) |
| 50duplex LEAD single biotin | GAGACCGAACGATCCTGTAATGTCCTAG CAAGCCAGAATTCGGCAGCGTCTTTTTT TTTTTTTTTTTTTTTTTTTTTTTTTTTT*T*T*T*T*T*T | Single biotin-modified dT in BOLD; the six dT nucelotides at the 3’ end are connected by phosphorothioate bonds (*) |
| 50duplex LEAD dual biotin | GAGACCGAACGATCCTGTAATGTCCTAG CAAGCCAGAATTCGGCAGCGTCTTTTTT TTTTTTTTTTTTTTTTTTTTTTTTTTTT*T*T*T*T*T*T | Two biotin-modified dT’s in BOLD; the six dT nucleotides at the 3’ end are connected by phosphorothioate bonds (*) |
| M.HpaII LAG | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTGACGCTGC-(5-F-dC)-GGATTCTGGCTTGCTAGGACATTACAGGATCGTTCGGTCTC | The position of a 5-fluorodeoxycytidine is indicated by (5-F-dC) |
| M.HpaII LEAD | GAGACCGAACGATCCTGTAATGTCCTAG CAAGCCAGAATCCGGCAGCGTCTTTTTT TTTTTTTTTTTTTTTTTTTTTTTTTTTT*T*T*T*T*T*T | The six dT nucleotides at the 3’ end are connected by phosphorothioate bonds (*) |





