Genetic epidemiology of dengue viruses in phase III trials of the CYD tetravalent dengue vaccine and implications for efficacy
Figures
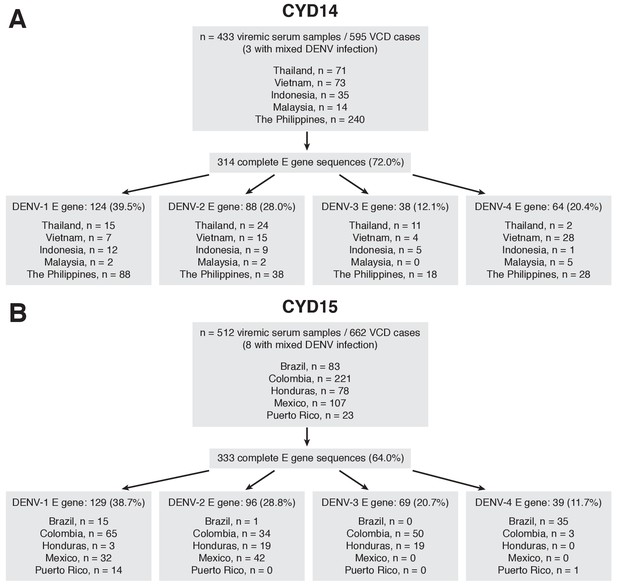
Sequencing flow chart for samples obtained in CYD-TDV trials.
(A) CYD14, (B) CYD15.
-
Figure 1—source data 1
Sequence alignment of DENV-1 prM and E genes from CYD-TDV trials.
- https://doi.org/10.7554/eLife.24196.005
-
Figure 1—source data 2
Sequence alignment of DENV-2 prM and E genes from CYD-TDV trials.
- https://doi.org/10.7554/eLife.24196.006
-
Figure 1—source data 3
Sequence alignment of DENV-3 prM and E genes from CYD-TDV trials.
- https://doi.org/10.7554/eLife.24196.007
-
Figure 1—source data 4
Sequence alignment of DENV-4 prM and E genes from CYD-TDV trials.
- https://doi.org/10.7554/eLife.24196.008
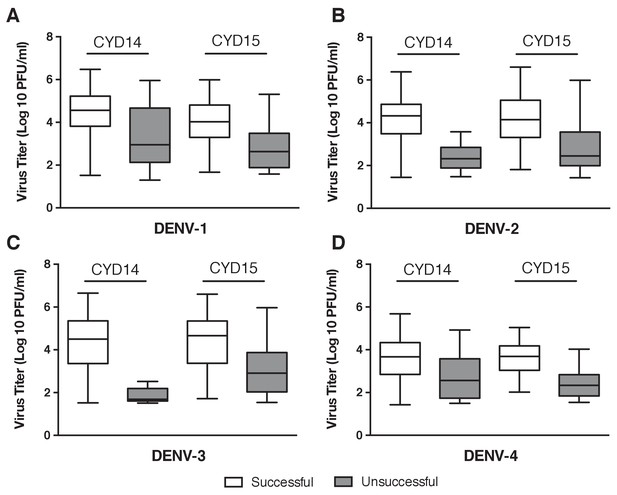
Probability of sequencing success versus viremia.
White boxes show the range and IQR of viremia levels from successful sequencing attempts, grey boxes show the range and IQR of viremia levels from unsuccessful sequencing attempts. (A) DENV-1, (B) DENV-2, (C) DENV-3, (D) DENV-4.
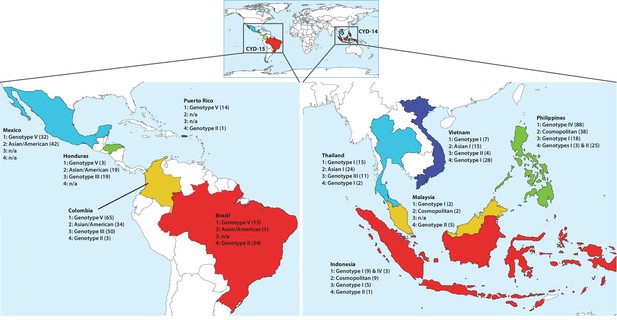
Distribution of DENV serotypes and genotypes sequenced in CYD14 and CYD15 vaccine trials by country.
Numbers in parentheses indicate the total number of samples of each genotype for which complete or partial E gene sequences were obtained.
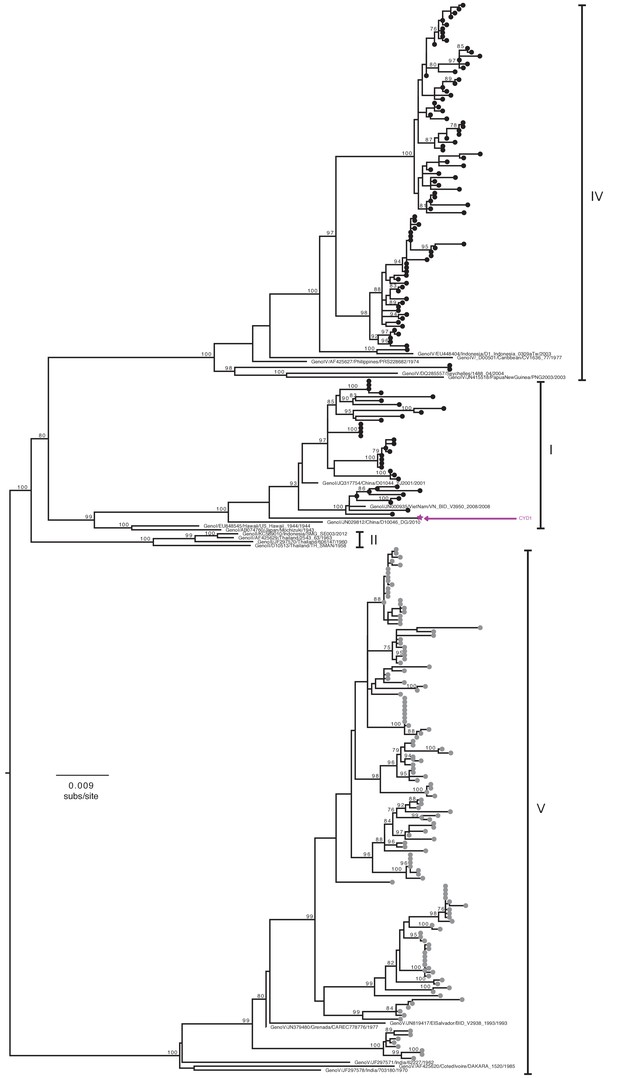
Genotype assignment of DENV-1 E gene sequences obtained from VCD samples in CYD14 and CYD15.
Phylogenetic trees showing the distribution of DENV-1 genotypes for which E genes were sequenced in CYD14 and CYD15 vaccine trials. E gene sequences were aligned with up to five publically available reference sequences of every known human DENV-1 genotype and a maximum likelihood phylogeny was constructed. Genotypes were assigned when sequences fell into a known genotype lineage with high bootstrap support. Bootstrap support values for nodes > 75% are indicated. Black dots at the tips indicate CYD14 sequences, grey dots indicate CYD15 sequences, and purple stars indicate the DENV CYD-TDV vaccine sequence for each serotype.
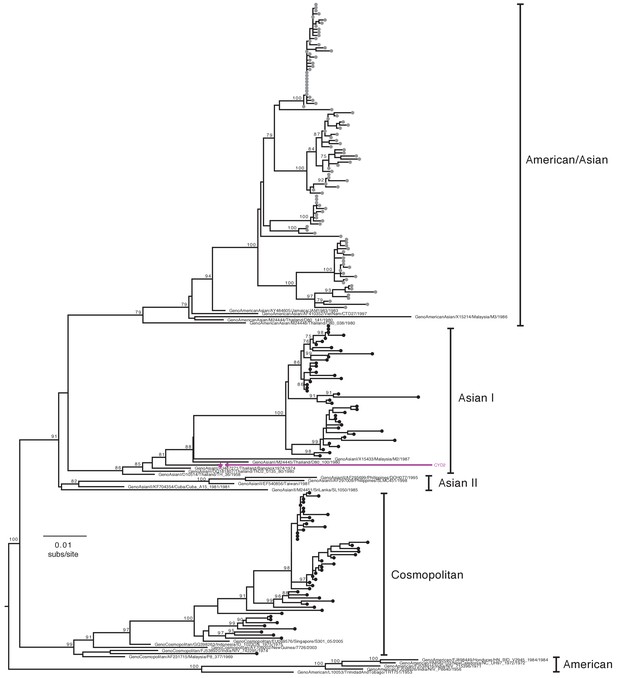
Genotype assignment of DENV-2 E gene sequences obtained from VCD samples in CYD14 and CYD15.
Phylogenetic trees showing the distribution of DENV-2 genotypes for which E genes were sequenced in CYD14 and CYD15 vaccine trials. E gene sequences were aligned with up to five publically available reference sequences of every known human DENV-2 genotype and a maximum likelihood phylogeny was constructed. Genotypes were assigned when sequences fell into a known genotype lineage with high bootstrap support. Bootstrap support values for nodes > 75% are indicated. Black dots at the tips indicate CYD14 sequences, grey dots indicate CYD15 sequences, and purple stars indicate the DENV CYD-TDV vaccine sequence for each serotype.
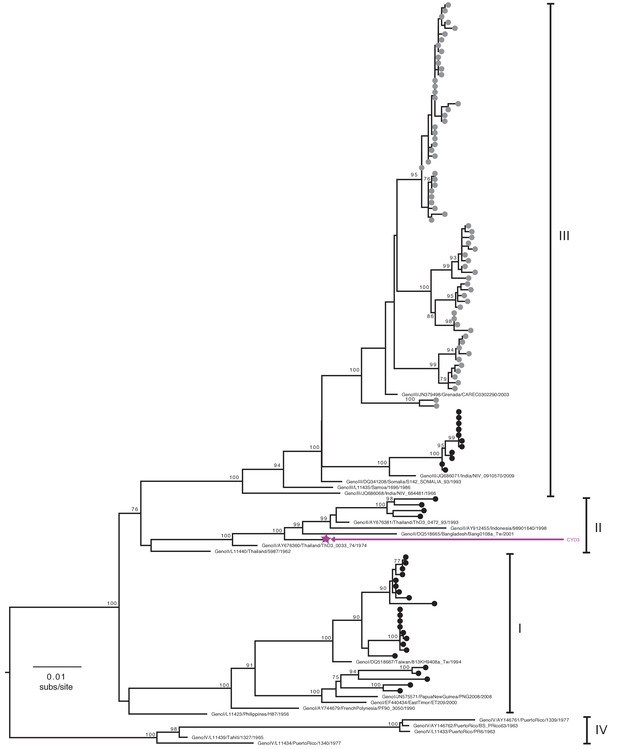
Genotype assignment of DENV-3 E gene sequences obtained from VCD samples in CYD14 and CYD15.
Phylogenetic trees showing the distribution of DENV-3 genotypes for which E genes were sequenced in CYD14 and CYD15 vaccine trials. E gene sequences were aligned with up to five publically available reference sequences of every known human DENV-3 genotype and a maximum likelihood phylogeny was constructed. Genotypes were assigned when sequences fell into a known genotype lineage with high bootstrap support. Bootstrap support values for nodes > 75% are indicated. Black dots at the tips indicate CYD14 sequences, grey dots indicate CYD15 sequences, and purple stars indicate the DENV CYD-TDV vaccine sequence for each serotype.
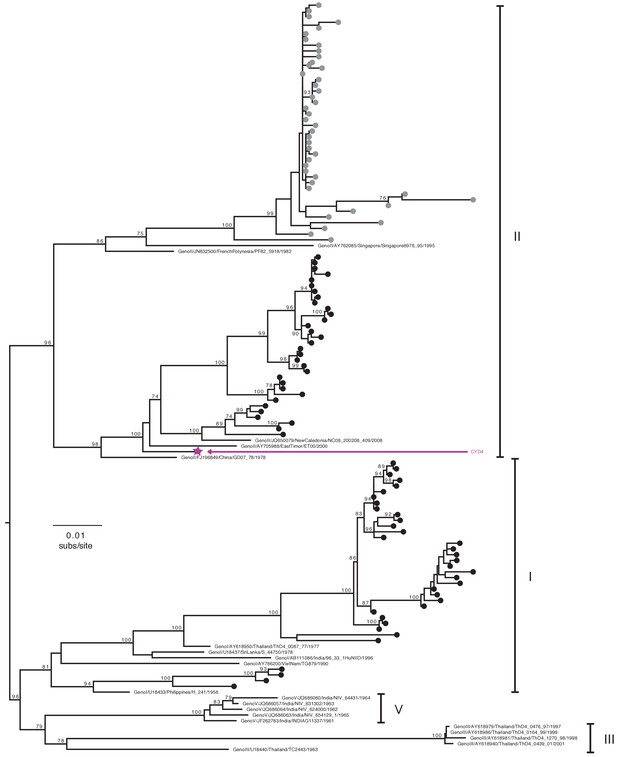
Genotype assignment of DENV-4 E gene sequences obtained from VCD samples in CYD14 and CYD15.
Phylogenetic trees showing the distribution of DENV-4 genotypes for which E genes were sequenced in CYD14 and CYD15 vaccine trials. E gene sequences were aligned with up to five publically available reference sequences of every known human DENV-5 genotype and a maximum likelihood phylogeny was constructed. Genotypes were assigned when sequences fell into a known genotype lineage with high bootstrap support. Bootstrap support values for nodes > 75% are indicated. Black dots at the tips indicate CYD14 sequences, grey dots indicate CYD15 sequences, and purple stars indicate the DENV CYD-TDV vaccine sequence for each serotype.
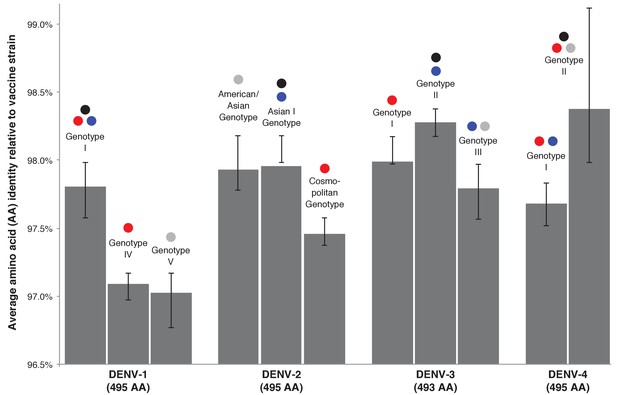
Average genotype-specific amino acid identity of DENV isolated in CYD-TDV trials compared to the vaccine strain of the corresponding DENV serotype.
Black bars indicate the IQR of the full sample set. Coloured dots show the geographic regions from which each genotype was collected – red: CYD14, maritime SE Asia; blue: CYD14, mainland SE Asia; grey: CYD15, Americas. Black dots indicate the genotype of the serotype-specific CYD-TDV vaccine component.
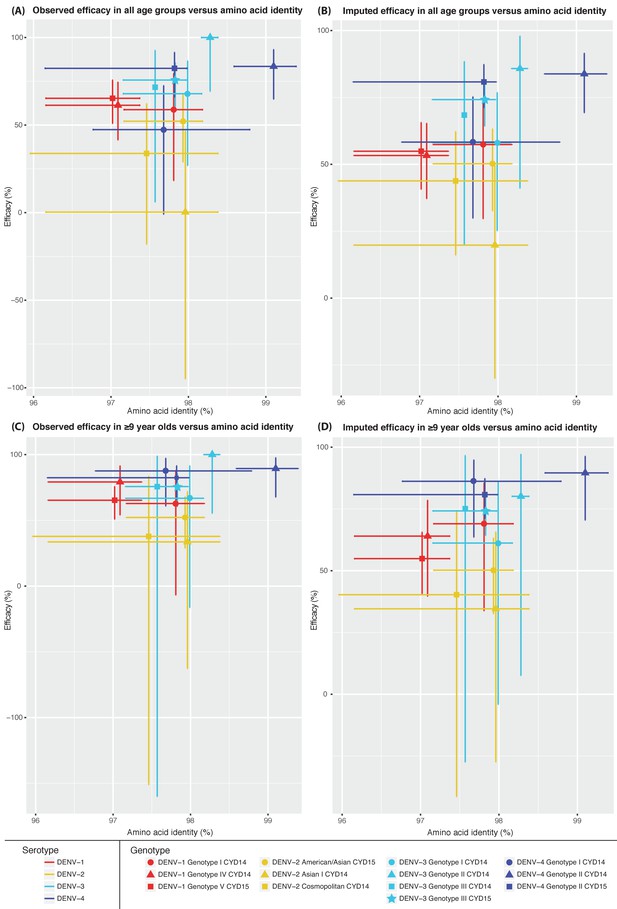
Genotype-specific amino acid identity of DENV isolated in CYD-TDV trials compared to the vaccine strain of the corresponding DENV serotype versus vaccine efficacy.
Symbols indicate the intersection of mean amino acid identity to CYD-TDV components and mean genotype-specific vaccine efficacy. Bars on the x-axis indicate the range of pairwise amino acid identities of DENV isolated in the trials compared to the CYD-TDV component. Bars on the y-axis indicate the 95% confidence intervals calculated for vaccine efficacy estimates. DENV-1 genotypes are shown in red, DENV-2 in yellow, DENV-3 in aqua, DENV-4 in blue. (A) Mean amino acid identity versus observed vaccine efficacy across all age groups. (B) Mean amino acid identity versus imputed vaccine efficacy across all age groups. (C) Mean amino acid identity versus observed vaccine efficacy in subjects ≥ 9 years of age. (D) Mean amino acid identity versus imputed vaccine efficacy in subjects ≥ 9 years of age.
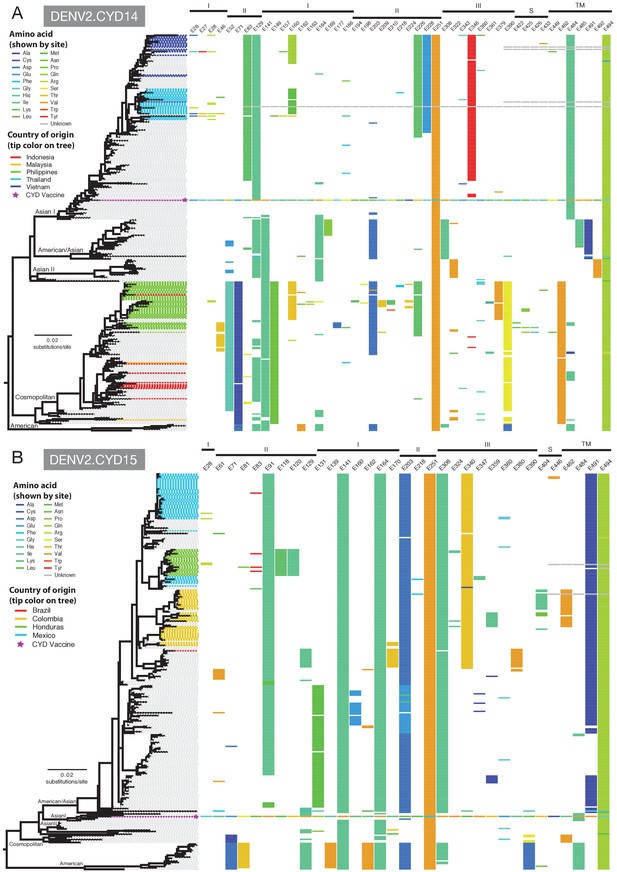
Amino acid differences between the DENV-2 E gene vaccine sequence, DENV-2 viruses isolated in CYD14 and CYD15 vaccine trials, and representative subsets of publically available DENV-2 sequences from the vaccine trial sites.
(A) CYD14 DENV-2 phylogeny, (B) CYD15 DENV-2 phylogeny. Coloured tips on the trees show sequences isolated in the CYD-TDV trials (country of origin coloured as indicated in the key) and the vaccine sequence (purple star); grey tips indicate publicly available sequences isolated from other studies in the countries of interest. Columns to the right indicate amino acid sites at which variation was observed in two or more CYD14/CYD15 sequences. Numbers at the top of columns indicate the amino acid site within the E gene. Bars at the top of the figures indicate the E gene domain of the site. Amino acids at variable sites in the E gene sequence of the vaccine component are shown in colour. For all other sequences, a lack of colour indicates an amino acid identical to that of the vaccine component at that site.
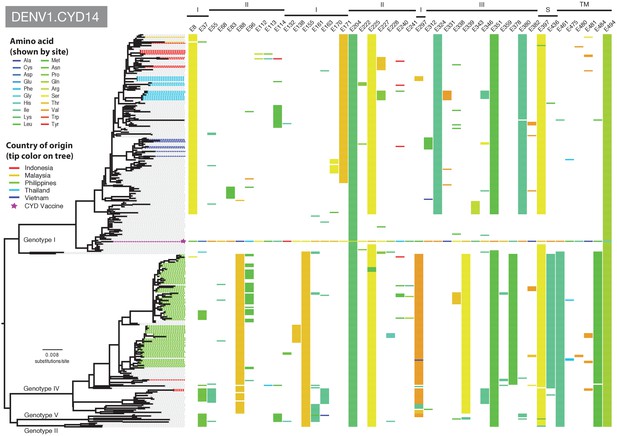
Amino acid differences between DENV-1 E gene vaccine sequences, DENV-1 viruses isolated in CYD14 vaccine trials, and representative subsets of publically available DENV-1 sequences from the vaccine trial sites.
Coloured tips on the trees show sequences isolated in the CYD14 trial (country of origin coloured as indicated in the key) and the respective vaccine sequence (purple star); grey tips indicate publicly available sequences isolated from other studies in the countries of interest. Columns to the right indicate amino acid sites at which variation was observed in two or more CYD14 sequences. Numbers at the top of columns indicate the amino acid site within the E gene. Bars at the top of the figures indicate the E gene domain of the site. Amino acids at variable sites in the E gene sequence of the vaccine component are shown in colour. For all other sequences, a lack of colour indicates an amino acid identical to that of the vaccine component at that site.
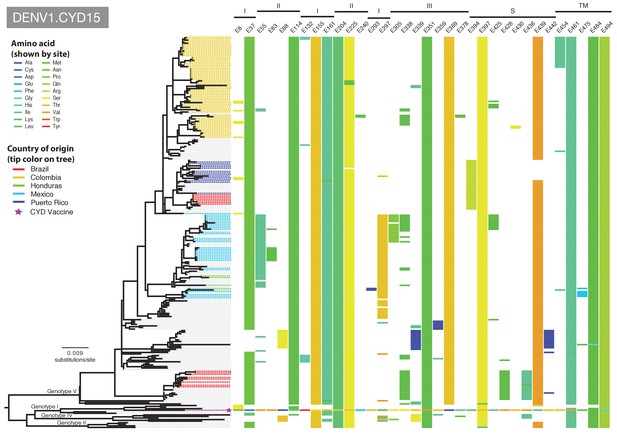
Amino acid differences between DENV-1 E gene vaccine sequences, DENV-1 viruses isolated in CYD15 vaccine trials, and representative subsets of publically available DENV-1 sequences from the vaccine trial sites.
Coloured tips on the trees show sequences isolated in the CYD15 trial (country of origin coloured as indicated in the key) and the respective vaccine sequence (purple star); grey tips indicate publicly available sequences isolated from other studies in the countries of interest. Columns to the right indicate amino acid sites at which variation was observed in two or more CYD15 sequences. Numbers at the top of columns indicate the amino acid site within the E gene. Bars at the top of the figures indicate the E gene domain of the site. Amino acids at variable sites in the E gene sequence of the vaccine component are shown in colour. For all other sequences, a lack of colour indicates an amino acid identical to that of the vaccine component at that site.
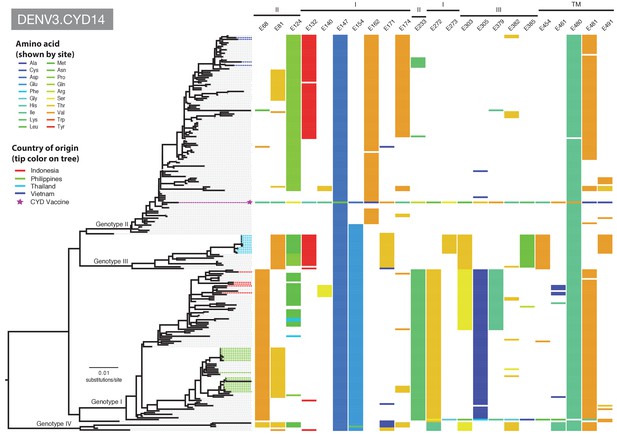
Amino acid differences between DENV-3 E gene vaccine sequences, DENV-3 viruses isolated in CYD14 vaccine trials, and representative subsets of publically available DENV-3 sequences from the vaccine trial sites.
Coloured tips on the trees show sequences isolated in the CYD14 trial (country of origin coloured as indicated in the key) and the respective vaccine sequence (purple star); grey tips indicate publicly available sequences isolated from other studies in the countries of interest. Columns to the right indicate amino acid sites at which variation was observed in two or more CYD14 sequences. Numbers at the top of columns indicate the amino acid site within the E gene. Bars at the top of the figures indicate the E gene domain of the site. Amino acids at variable sites in the E gene sequence of the vaccine component are shown in colour. For all other sequences, a lack of colour indicates an amino acid identical to that of the vaccine component at that site.
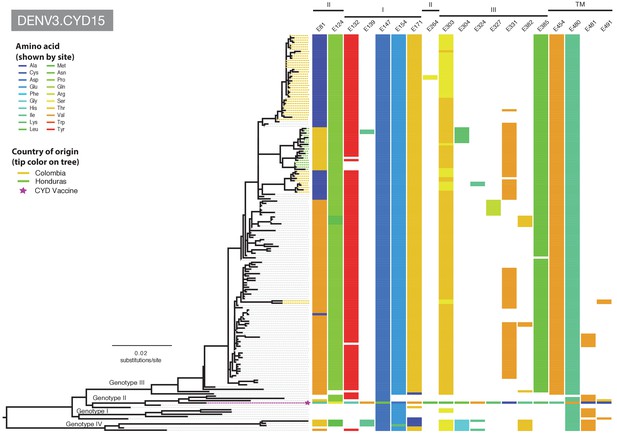
Amino acid differences between DENV-3 E gene vaccine sequences, DENV-3 viruses isolated in CYD15 vaccine trials, and representative subsets of publically available DENV-3 sequences from the vaccine trial sites.
Coloured tips on the trees show sequences isolated in the CYD15 trial (country of origin coloured as indicated in the key) and the respective vaccine sequence (purple star); grey tips indicate publicly available sequences isolated from other studies in the countries of interest. Columns to the right indicate amino acid sites at which variation was observed in two or more CYD15 sequences. Numbers at the top of columns indicate the amino acid site within the E gene. Bars at the top of the figures indicate the E gene domain of the site. Amino acids at variable sites in the E gene sequence of the vaccine component are shown in colour. For all other sequences, a lack of colour indicates an amino acid identical to that of the vaccine component at that site.
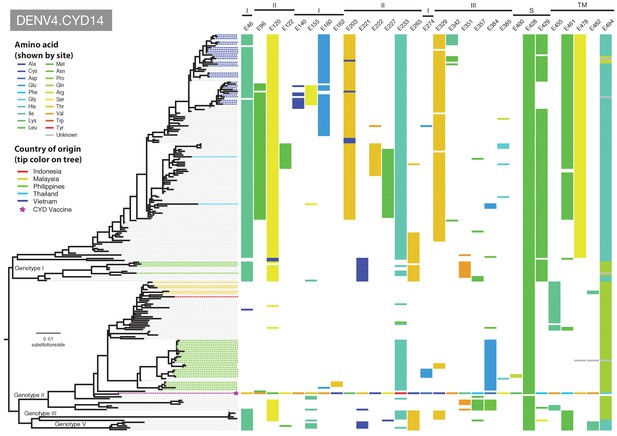
Amino acid differences between DENV-4 E gene vaccine sequences, DENV-4 viruses isolated in CYD14 vaccine trials, and representative subsets of publically available DENV-4 sequences from the vaccine trial sites.
Coloured tips on the trees show sequences isolated in the CYD14 trial (country of origin coloured as indicated in the key) and the respective vaccine sequence (purple star); grey tips indicate publicly available sequences isolated from other studies in the countries of interest. Columns to the right indicate amino acid sites at which variation was observed in two or more CYD14 sequences. Numbers at the top of columns indicate the amino acid site within the E gene. Bars at the top of the figures indicate the E gene domain of the site. Amino acids at variable sites in the E gene sequence of the vaccine component are shown in colour. For all other sequences, a lack of colour indicates an amino acid identical to that of the vaccine component at that site.
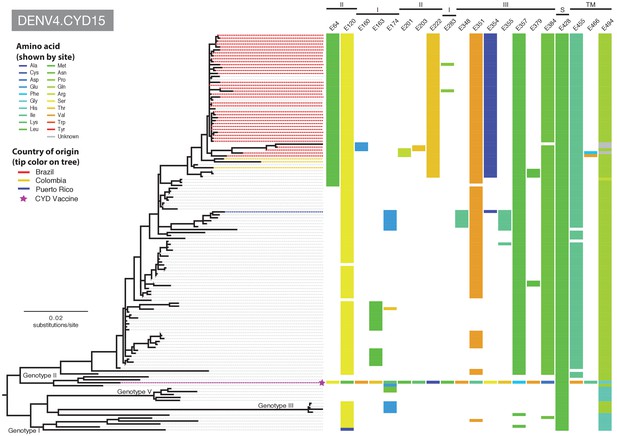
Amino acid differences between DENV-4 E gene vaccine sequences, DENV-4 viruses isolated in CYD15 vaccine trials, and representative subsets of publically available DENV-4 sequences from the vaccine trial sites.
Coloured tips on the trees show sequences isolated in the CYD15 trial (country of origin coloured as indicated in the key) and the respective vaccine sequence (purple star); grey tips indicate publicly available sequences isolated from other studies in the countries of interest. Columns to the right indicate amino acid sites at which variation was observed in two or more CYD15 sequences. Numbers at the top of columns indicate the amino acid site within the E gene. Bars at the top of the figures indicate the E gene domain of the site. Amino acids at variable sites in the E gene sequence of the vaccine component are shown in colour. For all other sequences, a lack of colour indicates an amino acid identical to that of the vaccine component at that site.
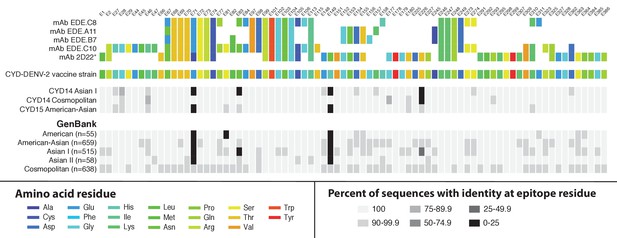
Sequence conservation between the DENV-2 vaccine component and wild-type DENV-2 viruses at epitope locations targeted by virus neutralising human mAbs.
Amino acid targets for five neutralising human mAbs (Fibriansah et al., 2015b; Rouvinski et al., 2015) are coloured as indicated in the key (top) and compared to the vaccine sequence and wild-type sequences obtained within the CYD14 and CYD15 trials (middle), as well as complete E gene sequences from wild-type DENV-2 available on GenBank (bottom). Sites are indicated at the top of columns. For wild-type virus populations, the darker the block, the greater the proportion of sequences with an amino acid differing from the target amino acid at that site. When disagreement between amino acids was observed between epitope targets (as at E67 and E71), wild-type sequences were compared to 2D22 as a reference, denoted by an asterisk.
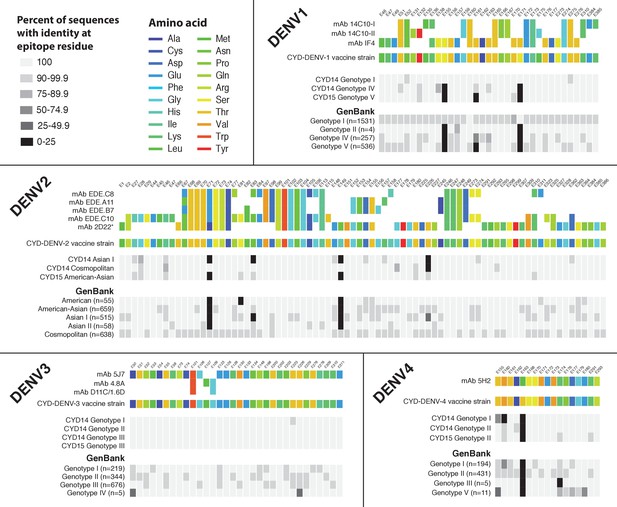
Sequence conservation between DENV vaccine components and wild-type DENV viruses at epitope locations targeted by virus neutralising human mAbs.
Amino acid targets for neutralising human mAbs (Fibriansah et al., 2014; Cockburn et al., 2012a; Fibriansah et al., 2015a, 2015b; Teoh et al., 2012; Rouvinski et al., 2015; Costin et al., 2013) are coloured as indicated in the key (top) and compared to the vaccine sequence and wild-type sequences obtained within the CYD14 and CYD15 trials (middle), as well as complete E gene sequences from wild-type DENV available on GenBank (bottom, numbers in parentheses indicate the number of sequences used for comparison). Sites are indicated at the top of columns. For wild-type virus populations, the darker the block, the greater the proportion of sequences with an amino acid differing from the target amino acid at that site.
Tables
Observed and imputed efficacy of CYD-TDV in all participants who received ≥1 injection (intention to treat) by serotype and genotype.
https://doi.org/10.7554/eLife.24196.025| CYD dengue vaccine group | Control group | Vaccine efficacy Observed | Vaccine Efficacy with imputation for missing genotype data | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Person-years at risk | Density incidence (95% CI) | Cases | Person-years at risk | Density incidence (95% CI) | % | (95% CI) | % | (95% CI) | ||
| Serotype 1 | 63.1 | (52.7; 71.2) | 54.7 | (45.4; 62.3) | |||||||
| Genotype I CYD14CYD | 15 | 13742 | 0.1 (0.1; 0.2) | 18 | 6796 | 0.3 (0.2; 0.4) | 58.8 | (18.3; 79.5) | 57.4 | (29.7; 74.2) | |
| Genotype IV CYD14 | 40 | 13742 | 0.3 (0.2; 0.4) | 51 | 6796 | 0.8 (0.6; 1.0) | 61.3 | (41.5; 74.5) | 53.3 | (37.2; 65.3) | |
| Genotype V CYD15 | 53 | 27016 | 0.2 (0.1; 0.3) | 76 | 13434 | 0.6 (0.4; 0.7) | 65.3 | (50.9; 75.7) | 54.9 | (40.7; 65.6) | |
| p-value* | 0.8614 | 0.9912 | |||||||||
| Serotype 2 | 39.1 | (18.9; 54.3) | 43.0 | (29.4; 53.9) | |||||||
| American/Asian CYD15 | 48 | 27035 | 0.2 (0.1; 0.2) | 50 | 13461 | 0.4 (0.3; 0.5) | 52.2 | (28.9; 67.9) | 50.2 | (32.6; 63.2) | |
| Asian I CYD14CYD | 28 | 13766 | 0.2 (0.1; 0.3) | 14 | 6856 | 0.2 (0.1; 0.3) | 0.3 | (−94.9; 46.6) | 19.8 | (−30.0; 49.6) | |
| Cosmopolitan CYD14 | 28 | 13766 | 0.2 (0.1; 0.3) | 21 | 6856 | 0.3 (0.2; 0.5) | 33.8 | (−18.0; 62.2) | 43.8 | (16.1; 62.2) | |
| p-value* | 0.1469 | 0.2493 | |||||||||
| Serotype 3 | 75.1 | (62.9; 83.3) | 71.6 | (63.0; 78.3) | |||||||
| Genotype I CYD14 | 9 | 13835 | <0.1 (0.0; 0.1) | 14 | 6895 | 0.2 (0.1; 0.3) | 67.9 | (26.9; 86.6) | 58.1 | (25.2; 76.8) | |
| Genotype II CYD14CYD | 0 | 13835 | 0.0 (0.0; 0.0) | 4 | 6895 | <0.1 (0.0; 0.1) | 100.0 | (69.3; 100.0) | 85.8 | (41.1; 97.9) | |
| Genotype III CYD14 | 4 | 13835 | <0.1 (0.0; 0.1) | 7 | 6895 | 0.1 (0.0; 0.2) | 71.6 | (6.1; 92.6) | 68.4 | (19.8; 88.4) | |
| Genotype III CYD15 | 23 | 27060 | <0.1 (0.1; 0.1) | 47 | 13459 | 0.3 (0.3; 0.5) | 75.7 | (60.5; 85.5) | 74.2 | (64.3; 81.4) | |
| Genotype III CYD14 + CYD15 | 27 | 40896 | <0.1 (0.0; 0.1) | 54 | 20354 | 0.3 (0.2; 0.3) | 75.2 | (61.0; 84.6) | 73.7 | (64.3; 80.8) | |
| p-value* | 0.3751 | 0.2561 | |||||||||
| Serotype 4 | 74.1 | (61.7; 82.5) | 76.9 | (69.5; 82.6) | |||||||
| Genotype I CYD14 | 19 | 13826 | 0.1 (0.1; 0.2) | 18 | 6874 | 0.3 (0.2; 0.4) | 47.4 | (−0.9; 72.5) | 58.3 | (29.9; 75.2) | |
| Genotype II CYD14CYD | 8 | 13826 | <0.1 (0.0; 0.1) | 24 | 6874 | 0.3 (0.2; 0.5) | 83.5 | (64.8; 93.1) | 83.8 | (69.3; 91.5) | |
| Genotype II CYD15CYD | 11 | 27063 | <0.1 (0.0; 0.1) | 31 | 13442 | 0.2 (0.2; 0.3) | 82.4 | (66.0; 91.5) | 80.8 | (71.2; 87.3) | |
| Genotype II CYD14 + CYD15CYD | 19 | 40890 | <0.1 (0.0; 0.1) | 55 | 20316 | 0.3 (0.2; 0.4) | 82.9 | (71.7; 90.1) | 81.8 | (74.3; 87.1) | |
| p-value* | 0.0072 | 0.0086 | |||||||||
-
Cases: number of subjects with at least one sequenced symptomatic virologically-confirmed dengue episode during the active phase of follow-up.
Density incidence: data indicate cases per 100 person-years at risk.
-
*The p-value was obtained by testing the heterogeneity of genotype distribution between groups (within each serotype) using a Chi2 (or Fisher’s exact test).
CYD Genotype of the serotype-specific CYD-TDV vaccine component.
Observed and imputed efficacy of CYD-TDV for subjects 9 years and older who received ≥1 injection (intention to treat) by serotype and genotype
https://doi.org/10.7554/eLife.24196.026| CYD dengue vaccine group | Control group | Vaccine efficacy Observed | Vaccine Efficacy with imputation for missing genotype data | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Person-years at risk | Density incidence (95% CI) | Cases | Person-years at risk | Density incidence (95% CI) | % | (95% CI) | % | (95% CI) | ||
| Serotype 1 | 67.7 | (56.1; 76.3) | 58.4 | (47.7; 66.9) | |||||||
| Genotype I CYD14CYD | 6 | 6683 | <0.1 (0.0; 0.2) | 8 | 3306 | 0.2 (0.1; 0.5) | 62.8 | (−6.8; 87.8) | 69.0 | (33.8; 85.5) | |
| Genotype IV CYD14 | 8 | 6683 | 0.1 (0.1; 0.2) | 19 | 3306 | 0.6 (0.3; 0.9) | 79.2 | (54.1; 91.4) | 64.0 | (39.7; 78.5) | |
| Genotype V CYD15 | 53 | 27016 | 0.2 (0.1; 0.3) | 76 | 13434 | 0.6 (0.4; 0.7) | 65.3 | (50.9; 75.7) | 54.9 | (40.7; 65.6) | |
| p-value* | 0.5213 | 0.5400 | |||||||||
| Serotype 2 | 48.6 | (27.4; 63.7) | 47.1 | (31.3; 59.2) | |||||||
| American/Asian CYD15 | 48 | 27035 | 0.2 (0.1; 0.2) | 50 | 13461 | 0.4 (0.3; 0.5) | 52.2 | (28.9; 67.9) | 50.2 | (32.6; 63.2) | |
| Asian I CYD14CYD | 12 | 6687 | 0.2 (0.1; 0.3) | 9 | 3330 | 0.3 (0.1; 0.5) | 33.6 | (−62.7; 71.9) | 34.6 | (−27.4; 65.7) | |
| Cosmopolitan CYD14 | 5 | 6687 | <0.1 (0.0; 0.2) | 4 | 3330 | 0.1 (0.0; 0.3) | 37.8 | (−151; 83.5) | 40.3 | (−41.4; 74.3) | |
| p-value* | 0.7736 | 0.7253 | |||||||||
| Serotype 3 | 76.0 | (62.3; 84.7) | 73.6 | (64.4; 80.4) | |||||||
| Genotype I CYD14 | 4 | 6715 | <0.1 (0.0; 0.2) | 6 | 3347 | 0.2 (0.1; 0.4) | 66.8 | (−16.3; 91.5) | 61.2 | (−4.1; 86.1) | |
| Genotype II CYD14CYD | 0 | 6715 | 0.0 (0.0; 0.1) | 3 | 3347 | <0.1 (0.0; 0.3) | 100.0 | (55.4; 100.0) | 80.1 | (7.6; 97.1) | |
| Genotype III CYD14 | 1 | 6715 | <0.1 (0.0; 0.1) | 2 | 3347 | <0.1 (0.0; 0.2) | 75.1 | (−160; 98.8) | 75.1 | (−27.4; 96.6) | |
| Genotype III CYD15 | 23 | 27060 | <0.1 (0.1; 0.1) | 47 | 13459 | 0.3 (0.3; 0.5) | 75.7 | (60.5; 85.5) | 74.2 | (64.3; 81.4) | |
| Genotype III CYD14 + CYD15 | 24 | 33775 | <0.1 (0.0; 0.1) | 49 | 16806 | 0.3 (0.2; 0.4) | 75.7 | (60.8; 85.3) | 74.3 | (64.7; 81.4) | |
| p-value* | 0.5928 | 0.6985 | |||||||||
| Serotype 4 | 85.2 | (74.6; 91.4) | 83.2 | (76.2; 88.2) | |||||||
| Genotype I CYD14 | 3 | 6716 | <0.1 (0.0; 0.1) | 12 | 3327 | 0.4 (0.2; 0.6) | 87.6 | (60.9; 97.2) | 86.2 | (63.6; 94.8) | |
| Genotype II CYD14CYD | 3 | 6716 | <0.1 (0.0; 0.1) | 14 | 3327 | 0.4 (0.2; 0.7) | 89.4 | (67.7; 97.6) | 89.6 | (70.5; 96.3) | |
| Genotype II CYD15CYD | 11 | 27063 | <0.1 (0.0; 0.1) | 31 | 13442 | 0.2 (0.2; 0.3) | 82.4 | (66.0; 91.5) | 80.8 | (71.2; 87.3) | |
| Genotype II CYD14 + CYD15CYD | 14 | 33779 | <0.1 (0.0; 0.1) | 45 | 16769 | 0.3 (0.2; 0.4) | 84.6 | (72.6; 91.8) | 82.6 | (74.7; 88.1) | |
| p-value* | 1.0000 | 0.6678 | |||||||||
-
Cases: number of subjects with at least one sequenced symptomatic virologically-confirmed dengue episode during the active phase of follow-up.
Density incidence: data indicate cases per 100 person-years at risk.
-
*The p-value was obtained by testing the heterogeneity of genotype distribution between groups (within each serotype) using a Chi2 (or Fisher’s exact test).
CYD Genotype of the serotype-specific CYD-TDV vaccine component.
Estimation of the interaction between genotype and vaccine group for symptomatic VCD detected during the active phase of follow-up by serotype in all participants who received >= 1 injection (intention to treat) (CYD14/CYD15).
The estimate of the interaction term between genotype and vaccine group is derived from Cox proportional hazards regression models including the vaccine group, the genotype and the interaction.
| Estimated interaction with observed vaccine efficacy | Estimated interaction with vaccine efficacy with imputation | ||||
|---|---|---|---|---|---|
| Serotype | Parameter | Parameter estimate | 95% | Parameter estimate | 95% |
| Serotype 1 | Genotype IV vs Genotype I | −0.058 | [−0.858; 0.743] | 0.095 | [−0.475; 0.665] |
| Genotype V vs Genotype I | −0.167 | [−0.936; 0.603] | 0.067 | [−0.492; 0.625] | |
| Serotype 2 | American/Asian vs Asian I | −0.732 | [−1.486; 0.022] | −0.471 | [−1.032; 0.089] |
| Cosmopolitan vs Asian I | −0.404 | [−1.259; 0.451] | −0.344 | [−0.966; 0.267] | |
| Serotype 3 | Genotype II vs Genotype I | −12.748 | [−729.203; 703.707] | −1.079 | [−2.754; 0.596] |
| Genotype III vs Genotype I | −0.251 | [−1.208; 0.705] | −0.459 | [−1.116; 0.198] | |
| Serotype 4 | Genotype II vs Genotype I | −1.114 | [−1.943; −0.285] | −0.8184 | [−1.434; −0.203] |
Estimation of the interaction between genotype and vaccine group for symptomatic VCD detected during the active phase of follow-up by serotype in subjects older than 9 years of age who received >= 1 injection (intention to treat) (CYD14/CYD15).
The estimate of the interaction term between genotype and vaccine group is derived from Cox proportional hazards regression models including the vaccine group, the genotype and the interaction.
| Estimated interaction with observed vaccine efficacy | Estimated interaction with vaccine efficacy with imputation | ||||
|---|---|---|---|---|---|
| Serotype | Parameter | Parameter estimate | 95% | Parameter estimate | 95% |
| Serotype 1 | Genotype IV vs Genotype I | −0.574 | [−1.917; 0.768] | 0.153 | [−0.760; 1.066] |
| Genotype V vs Genotype I | −0.061 | [−1.177; 1.054] | 0.385 | [−0.416; 1.186] | |
| Serotype 2 | American/Asian vs Asian I | −0.327 | [−1.277; 0.624] | −0.270 | [−0.987; 0.448] |
| Cosmopolitan vs Asian I | −0.064 | [−1.637; 1.510] | −0.089 | [−1.151; 0.972] | |
| Serotype 3 | Genotype II vs Genotype I | −13.019 | [−943.634; 917.597] | −0.664 | [−2.578; 1.250] |
| Genotype III vs Genotype I | −0.309 | [−1.665; 1.047] | −0.405 | [−1.443; 0.633] | |
| Serotype 4 | Genotype II vs Genotype I | 0.226 | [−1.174; 1.626] | 0.244 | [−0.789; 1.277] |
Additional files
-
Supplementary file 1
(a) Sequencing success rates in samples from virologically confirmed dengue cases in vaccine and control groups from the CYD-TDV trials.
(b) Mean percent identity between E gene amino acid sequences of the relevant serotype-specific CYD-TDV vaccine strain and virus populations sampled in CYD14/15. (c) Number of E gene sequences per genotype per country in CYD-TDV trials versus publicly available sequences on GenBank. I. CYD14, II. CYD15. (1d) Variation in the number of cases, imputed versus observed.
- https://doi.org/10.7554/eLife.24196.029
-
Supplementary file 2
Observed and imputed efficacy of CYD-TDV for subjects less than 9 years of age who received ≥1 injection (intention to treat) by serotype and genotype.
- https://doi.org/10.7554/eLife.24196.030
-
Supplementary file 3
Phred scores indicating sequence quality for all CYD14/15 DENV prM/E sequences.
- https://doi.org/10.7554/eLife.24196.031
-
Transparent reporting form
- https://doi.org/10.7554/eLife.24196.032





