Arabidopsis 14-3-3 epsilon members contribute to polarity of PIN auxin carrier and auxin transport-related development
Figures
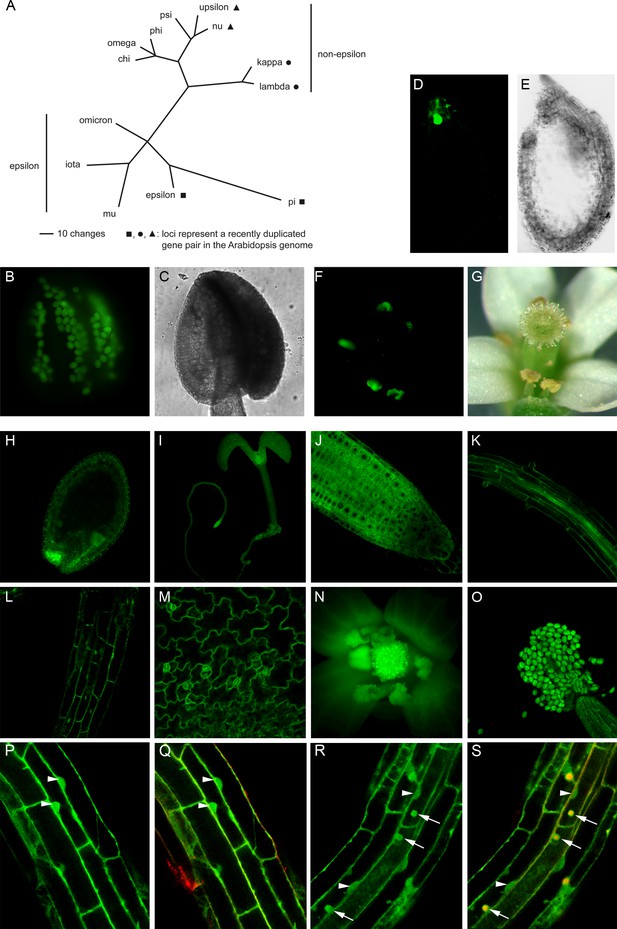
Phylogenetic tree of the Arabidopsis 14-3-3 isoforms and expression of genomic 14-3-3:GFP fusions under the control of the endogenous promoter.
(A) Unrooted phylogenetic tree of the Arabidopsis 14-3-3 protein family. Maximum parsimony analyses were performed using PAUP 4.0b10 (Altivec) with the bootstrap-algorithm and 1000 replica. (B, C) Expression of iota-GFP is restricted to pollen. (D) to (G) Pi-GFP is exclusively expressed in the chalazal cyst of the seed (D, E) and the connective tissue of the anthers (F, G). (H) to (O) The expression of nu-GFP is ubiquitous. Expression in seeds (H), seedlings (I), root tips (J), roots (K), hypocotyl (L), leaf epidermis (M) and floral organs (N, O) is shown. (P) to (S) Mu-GFP (P, R) and merge with FM4-64 (Q, S) in epidermal root cells (elongation zone) of seedlings (4d) treated with CHX (50 μM) for 60 min followed by treatment with CHX (P, Q) or CHX and 50 μM BFA (R, S) for 60 min. Note that mu-GFP localizes to both nucleus and cytoplasm but accumulates in BFA bodies upon inhibitor treatment. Arrowheads indicate nuclei, arrows mark BFA bodies.
-
Figure 1—source data 1
Expression patterns of 14-3-3 isoforms in various tissues of six plant species based on publicly accessible RNA-seq or microarray data.
Normalized data for the expression of 14-3-3 isoforms (absolute signal intensities) were obtained from the eFP Browser of the Bio-Analytic Resource for Plant Biology (BAR) (http://bar.utoronto.ca) (microarray data: A. thaliana, M. truncatula, P. trichocarpa, O. sativa, P. patens) or the Tomato Functional Genomics Database (http://ted.bti.cornell.edu/cgi-bin/TFGD/digital/home.cgi) (RNA-seq data: S. lycopersicum). Expression of ubiquitin is depicted for comparison. Individual Excel sheets have been created for the different plant species. Each Excel sheet lists detailed information of 14-3-3s, such as gene ID and phylogenetic subgroup as well as the origin of expression data including references.
- https://doi.org/10.7554/eLife.24336.003
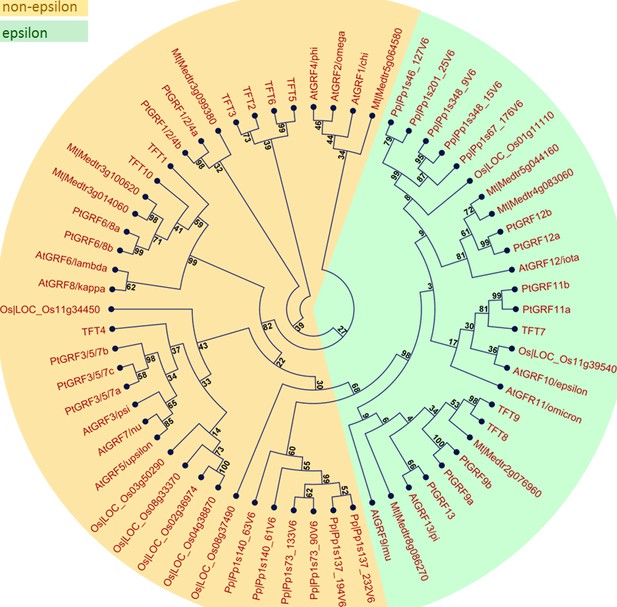
Phylogenetic relationship of 14-3-3 family members from six plant species.
Multiple alignment of 14-3-3 isoforms from A. thaliana (At), S. lycopersicum (TFT), M. truncatula (Mt), P. trichocarpa (Pt), O. sativa (Os), and P. patens (Pp) was performed using CLC Main Workbench 7.8. A maximum likelihood phylogenetic tree was constructed with 10,000 bootstrap replicates. Bootstrap support values are shown on each node. The two major groups are marked with different background colors. Detail information of 14-3-3s from the six plant species are listed in Figure 1.
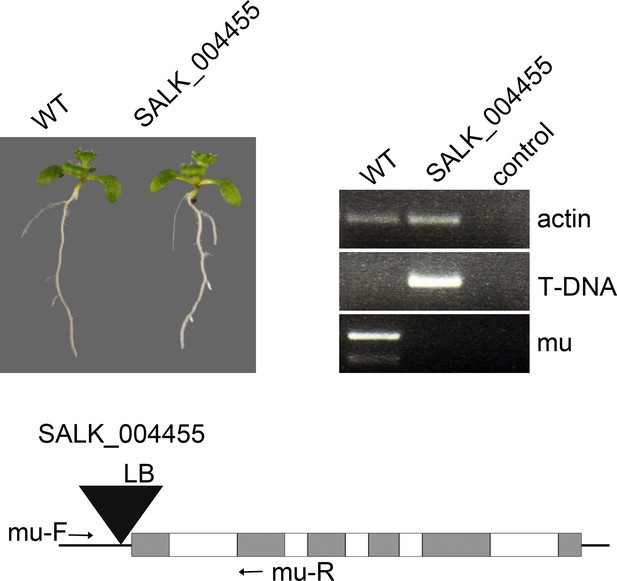
Phenotype of the homozygous 14-3-3 mu T-DNA allele (SALK_004455) under continuous light.
The primers indicated in the schematic representation of the T-DNA insertion (lower panel) were used to identify homozygous plants. The control PCR reactions contain no input genomic DNA.
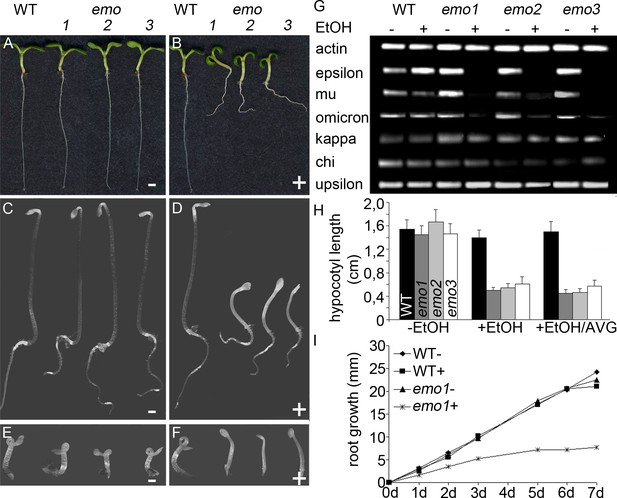
Ethanol-inducible emo-RNAi causes growth retardation phenotypes.
(A) to (F) Seedlings (wildtype and three independent emo-RNAi lines) grown either for 6 days in the light (A, B) or for 4 days in the dark (C–F) on noninductive (A, C, E) or inductive (0.1% (v/v) EtOH) medium (B, D, F), optionally supplemented with ACC (10 μM) (E, F). (G) Semiquantitative RT-PCR analysis of the transcript level of selected 14-3-3 isoforms in light-grown seedlings. (H) Hypocotyl length of etiolated seedlings (n = 30). (I) Primary root growth of seedlings grown for 4 days in the absence of ethanol followed by transfer to non-inductive (−) or inductive (+) medium for the indicated times (n = 16).
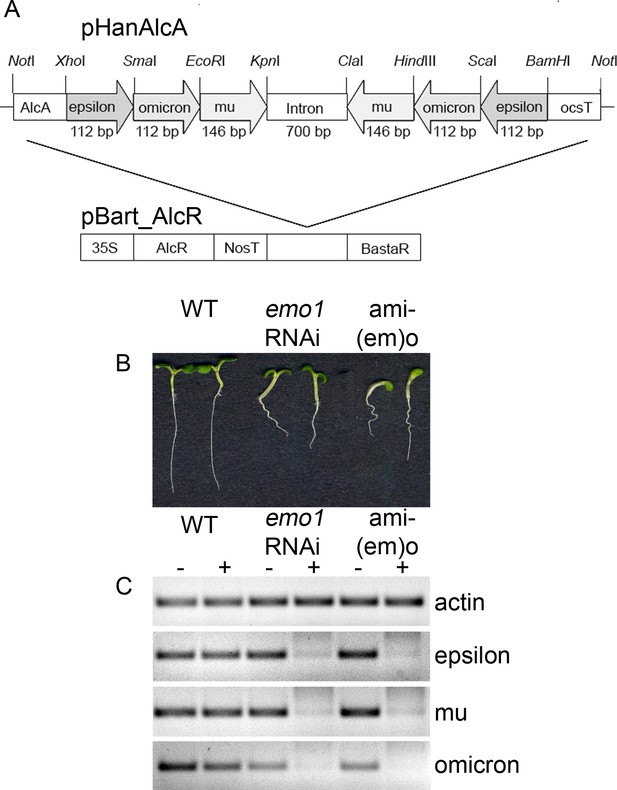
Ethanol-inducible amiRNA-(em)o causes growth retardation phenotypes.
(A) Schematic representation of the RNAi construct generated to reduce the expression of the 14-3-3 isoforms epsilon, mu and omicron. (B) Seedlings (two at a time, from left to right: wildtype, emo1-RNAi and amiRNA-(em)o)) grown for 6 days in the presence of ethanol. (C) Semiquantitative RT-PCR analysis of the transcript level of the targeted 14-3-3 isoforms in wildtype, emo1-RNAi and amiRNA-(em)o grown for 6 days in the absence (−) or presence (+) of ethanol.
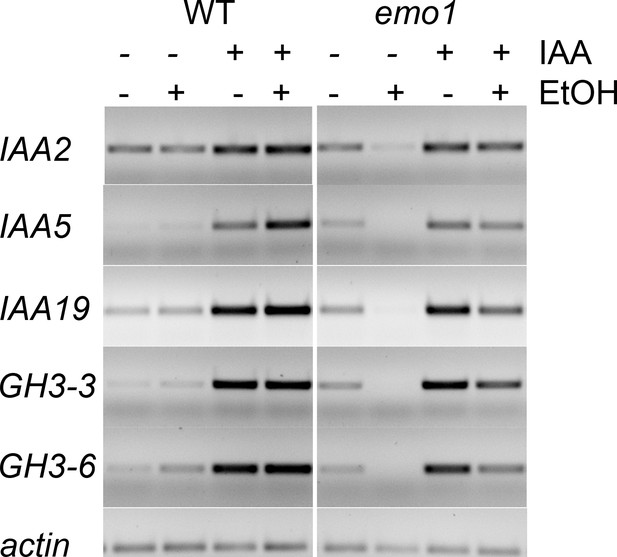
Auxin-induced gene expression is not compromised in emo-RNAi roots.
Semiquantitative RT-PCR analysis of the transcript level of selected auxin-induced genes in wildtype and emo1-RNAi seedlings grown for 7 days on MS medium followed by transfer to ethanol-containing (+) or -lacking (−) medium for 2 days and finally treated with mock (−) or 50 μM IAA (+) for 2 hr. Total RNA was extracted from root tissue.
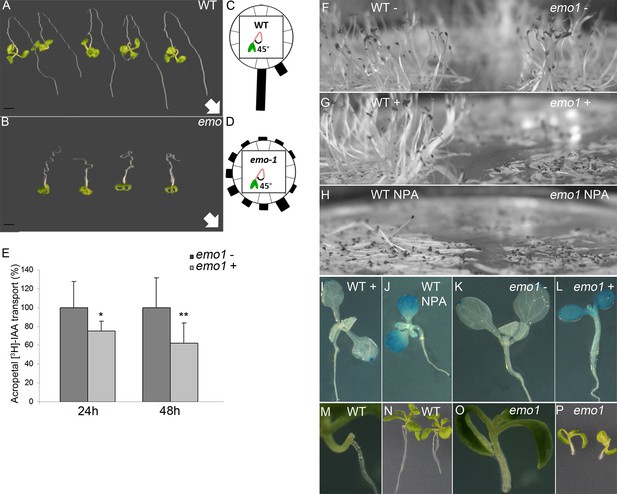
Ethanol-inducible emo-RNAi causes defects in the gravitropic growth response and auxin transport in both roots and aerial tissues.
(A) to (D) Gravitropic root growth response of emo1-RNAi (B) as compared to wildtype (A). Seedlings grown for 3 days in the absence of ethanol were transferred to inductive medium for 2 days, followed by reorientation (135°, arrows indicate gravity vector). Photos were taken after 5 days. Re-alignment to the gravity vector was recorded after 4 days (C, D, n = 30). (E) Acropetal auxin transport in emo1-RNAi roots. Seedlings grown for 4 days in the absence of ethanol were transferred to non-inductive (dark-gray bars) or inductive medium (light-gray bars) for 24 hr and 48 hr, respectively, followed by the application of agar droplets supplemented with radioactive IAA. Transport to the root tip was measured after 18 hr (n = 10 roots). (F) to (H) Etiolated wild-type (left side) and emo1-RNAi seedlings (right side) grown for 4 days in the absence (F, H) or presence of ethanol (G) and NPA (H). (I) to (L) Auxin response gradients visualized by DR5::GUS in wild type (I, J) and emo1-RNAi (K, L) in the absence (J, K) or presence of ethanol (I, L) and NPA (J). (M) to (P) Formation of adventitious roots in wild type (M, N) and emo1-RNAi (O, P) after 48 hr of ethanol induction followed by surgical removal of the primary root. Root regeneration 8 days after cutting is shown.
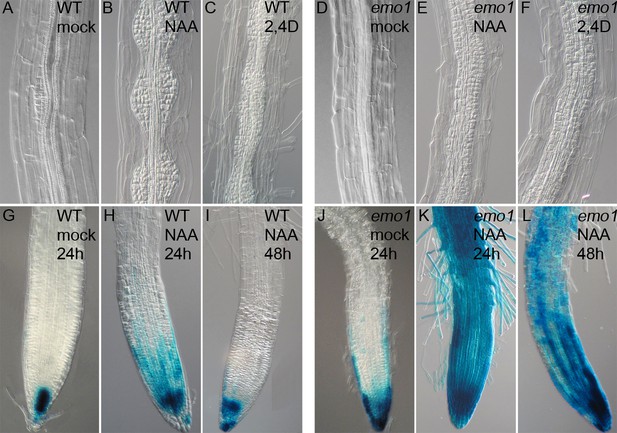
Disorganized lateral root primordia and failure in the establishment of auxin response gradients caused by emo-RNAi induction.
Seedlings grown for 4 days in the absence of ethanol were transferred to inductive medium for 24 hr followed by treatment with exogenous auxin.(A) to (F) Lateral root primordia of wildtype (A–C) and emo1-RNAi (D–F) after 48 hr of treatment with either mock solution (A, D), 1 μM NAA (B, E) or 1 μM 2,4D (C, F) in the presence of ethanol. (G) to (L) Auxin response gradients visualized by DR5::GUS in wild type (G–I) and emo1-RNAi (J–L) after treatment with either mock solution (G, J) or 0.1 μM NAA for either 24 hr (H, K) or 48 hr (I, L) in the presence of ethanol.
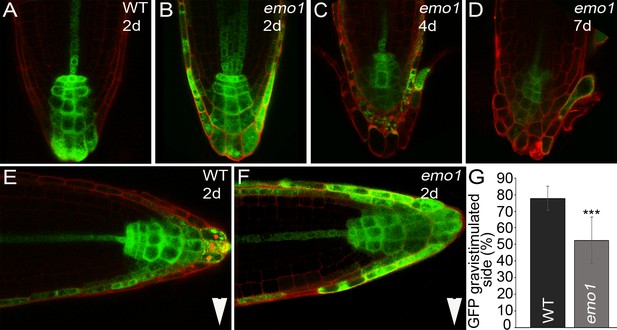
Changes in auxin distribution caused by emo-RNAi induction.
(A, B, E–G) DR5rev::GFP activity in root tips of wild type (A, E) and emo1-RNAi (B, F) after transfer to inductive medium for 2 days (A, B) and gravistimulation (6 hr, 90°) (E, F). Arrowheads indicate gravity vector. Percentage of GFP signal in the gravistimulated side of the root is shown in (G) (n = 10 roots). (C, D) DR5rev::GFP activity in root tips of emo1-RNAi after transfer to inductive medium for 4 days (C) and 7 days (D).
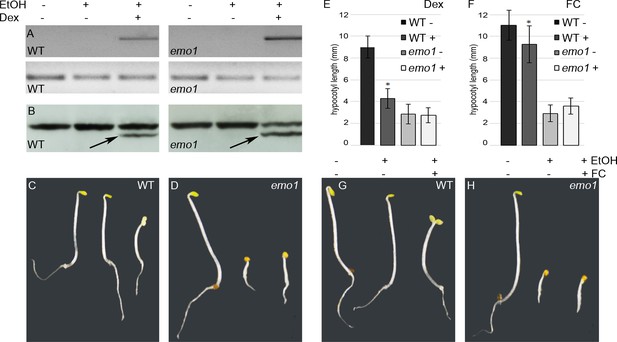
Dexamethasone-dependent expression of a constitutively active version of AHA2 does not rescue the emo-RNAi phenotype.
(A) to (E) Wild-type and emo1-RNAi seedlings were grown for 4 days in the dark in the absence or presence of ethanol and Dex (10 μM). Semiquantitative RT-PCR analysis of the transcript level of the C-terminally deleted AHA2 (A, upper panel, lower panel shows actin) and immunodetection of the H+-ATPase in microsomal membranes (B, arrows indicate the truncated version of the H+-pump) confirmed Dex-dependent expression of the transgene. Seedling growth (C, D) and measurement of the hypocotyl length (E, n = 30) is shown. (F) to (H) Treatment with fusicoccin (FC), an activator of the H+-ATPase, mimicked the effects observed in transgenic wild type upon Dex-treatment (see (C): shorter hypocotyls, absence of an apical hook and open cotyledons). Seedling growth (G, H) and measurement of the hypocotyl length (F, n = 30) in the absence or presence of FC is shown.
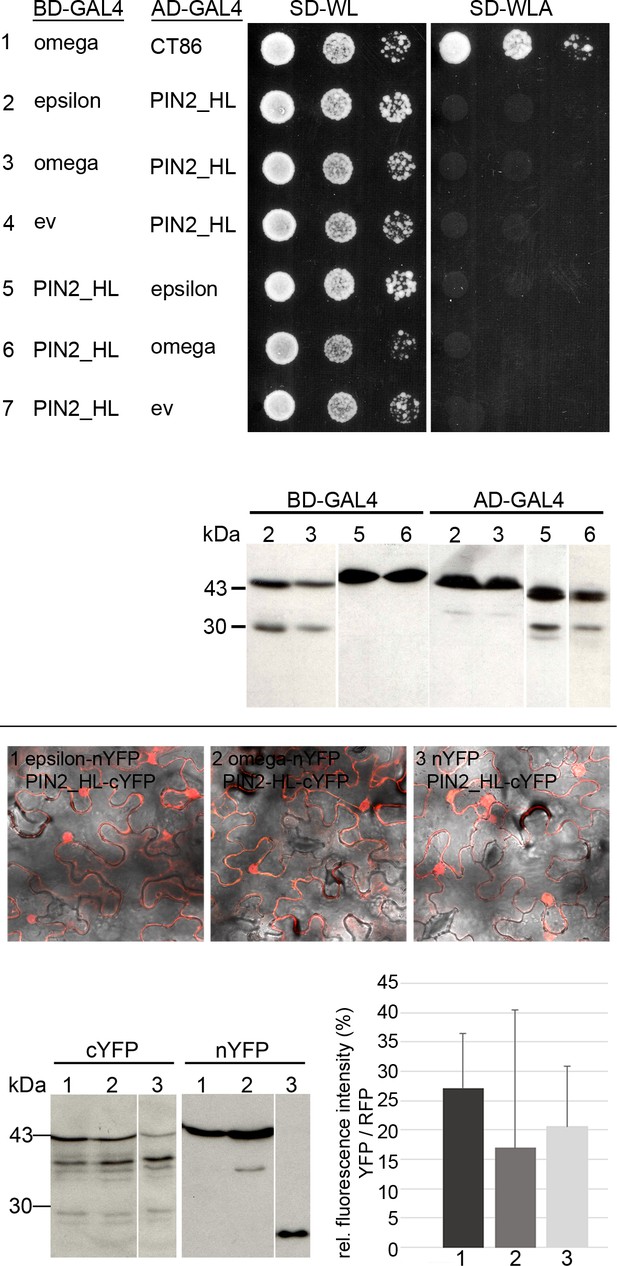
Yeast two-hybrid analysis and in planta interaction studies do not point to a direct interaction of 14-3-3 s with the PIN2 hydrophilic loop.
Upper panel: Yeast two-hybrid interaction analysis of the 14-3-3 isoforms epsilon or omega with the PIN2 hydrophilic loop region (top). As a positive control, the interaction of omega with the C-terminal domain of the H+-ATPase (CT86) is shown. Expression of the individual fusion proteins was confirmed by immunodetection (bottom). ev: empty vector. Lower panel: Ratiometric bimolecular fluorescence complementation analysis of the interaction of 14-3-3 epsilon or 14-3-3 omega with the PIN2 hydrophilic loop. Exemplary CLSM images (merge) of abaxial leaf epidermis cells of N. benthamiana 2 days after infiltration with A. tumefaciens containing pBiFCt-2in1-CC with the indicated cDNAs are shown (top). Expression of the individual fusion proteins was confirmed by immunodetection (bottom left). Quantification of the mean YFP fluorescence ratioed against RFP is shown (bottom right).
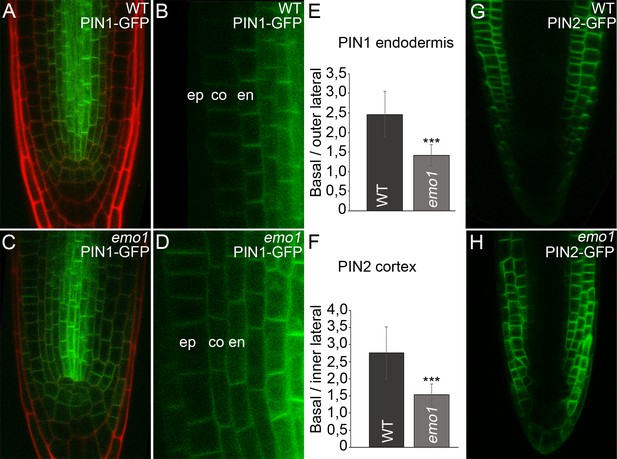
Ethanol-inducible emo-RNAi causes misexpression of PIN proteins in root tips.
(A) to (H) Expression of PIN1-GFP (A–D) and PIN2-GFP (G, H) in wildtype (A, B, G) and emo1-RNAi (C, D, H) seedlings grown for 4 days on inductive medium. The ratio of GFP intensity on the basal to that on the lateral side of the plasma membrane is shown (E, n ≥ 64 cells, 12 roots, F, n ≥ 75 cells, 10 roots). en: endodermis, co: cortex, ep: epidermis.
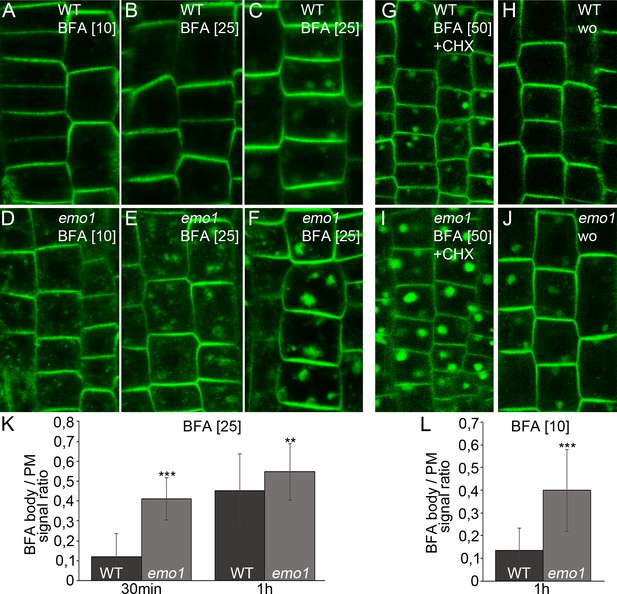
Ethanol-inducible emo-RNAi alters membrane trafficking processes.
(A) to (F) PIN2-GFP in epidermal root cells of wild-type (A–C) and emo1-RNAi (D–F) seedlings grown for 4 days on inductive medium and treated with BFA for 10 min (A, D), 30 min (B, E) or 60 min (C, F). The BFA concentration used (μM) is given in brackets. (G) to (J) PIN2-GFP in epidermal root cells of induced (4d) wild-type (G, H) and emo1-RNAi (I, J) seedlings treated with cycloheximide (CHX, 50 μM) for 60 min followed by concomitant treatment with CHX and 50 μM BFA (G, I) and subsequent washout of BFA (H, J) for 60 min (wo), respectively. (K, L) Ratio of PIN2-GFP intensity in BFA bodies and the PM (1 BFA body and 1 PM per cell, K: n ≥ 90 cells, 15 roots; L: n ≥ 75 cells, 11 roots).
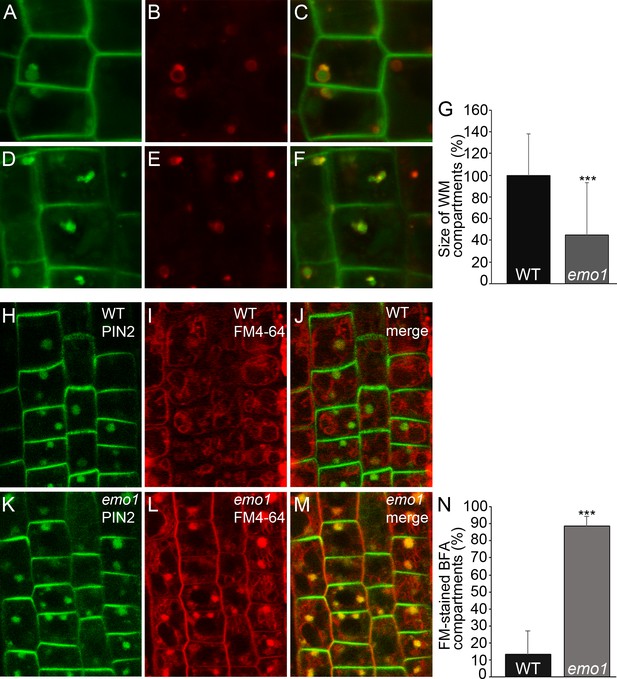
Ethanol-inducible emo-RNAi causes defects in wortmannin-compartment formation and endocytic transport to the vacuole.
(A) to (G) PIN2-GFP (A, D) and mRFP-ARA7 (B, E) in epidermal root cells of induced (4d) wildtype (A–C) and emo1-RNAi (D–F) seedlings treated with cycloheximide (CHX, 50 μM) for 60 min followed by concomitant treatment with CHX and WM (15 μM) for 120 min. Measurement of the relative size of WM-compartments is shown in (G) (n = 100). (H) to (N) PIN2-GFP (H, K) and FM4-64 (I, L) in epidermal root cells of induced (4d) wild-type (H–J) and emo1-RNAi (K–M) seedlings pulse labeled by FM4-64 (5 min) and subsequently treated with CHX (50 μM, 1 hr) followed by concomitant treatment with CHX and 50 μM BFA for 1 hr. Percentage of FM4-64 stained BFA bodies (J, M) is shown in (N) (n ≥ 115 cells, 10 roots).
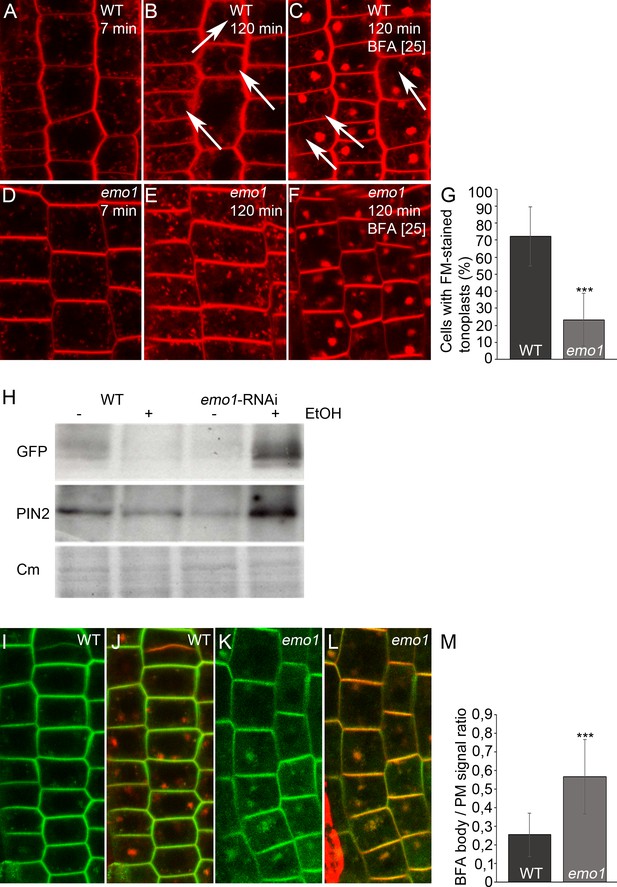
Ethanol-inducible emo RNAi delays endocytic trafficking from TGN/EE to the vacuole and alters endosome homeostasis.
(A) to (G) Root epidermal cells of induced (4d) wild-type (A–C) and emo1-RNAi (D–F) seedlings were stained with the endocytic tracer FM4-64 in the absence (A, B, D, E) or presence of 25 μM BFA (C, F) for the indicated times. Arrows indicate tonoplast staining. Percentage of cells exhibiting FM4-64-stained tonoplasts in the absence of BFA (see B, E) is shown in (G) (n ≥ 445 cells, 20 roots). (H) Immunodetection of PIN2-GFP in microsomal membranes prepared from wild-type and emo1-RNAi seedling roots grown in the absence or presence of ethanol (upper panel: GFP antibody, middle panel: PIN2 antibody). Coomassie staining (Cm) served as loading control (lower panel). (I) to (M) PIN2-GFP (I, K) and merge with FM4-64 (J, L) in epidermal root cells of induced (4d) wild-type (I, J) and emo1-RNAi (K, L) seedlings pretreated with FM4-64 (5 min) prior to concomitant treatment with 10 μM NAA and CHX for 30 min and followed by treatment with NAA, CHX and 50 μM BFA (90 min). The ratio of PIN2-GFP intensity in BFA bodies and the PM is shown in (M) (1 BFA body and 1 PM per cell, n ≥ 70 cells, 10 roots).
Tables
Analysis of 14-3-3 epsilon-GFP immunoprecipitates via mass spectrometry (MS) based on two biological replicates. This table lists only proteins with a possible role in membrane trafficking. Proteins depicted in a clustered manner (without blank line) represent alternative possibilities based on the MS-identified peptides.
| AGI code | Gene name | Description |
|---|---|---|
| At1g08680 | AGD14 | ADP-ribosylation factor (ARF) GTPase-activating protein |
| At1g09630 | RAB-A2a | Member of the RAB-A subfamily of small Rab GTPases |
| At1g16920 | RAB-A1b/BEX5 | |
| At3g15060 | RAB-A1g | |
| At4g18800 | RAB-A1d | |
| At5g45750 | RAB-A1c | |
| At5g60860 | RAB-A1f | |
| At1g12360 | KEULE/SEC11 | SNARE-interacting protein Sec1 protein |
| At1g14670 | Endomembrane protein 70 protein family | |
| At2g01970 | Endomembrane protein 70 protein family | |
| At5g37310 | ||
| At2g20790 | AP5M | AP-5 complex subunit mu |
| At2g25430 | ENTH/ANTH/VHS superfamily protein | |
| At2g37550 | AGD7 | ARF GTPase-activating protein AGD7 |
| At2g43160 | EPSIN2 | ENTH/ANTH/VHS superfamily protein |
| At3g59290 | EPSIN3 | |
| At3g09900 | RAB-E1e | Member of the RAB-E subfamily of small Rab GTPases |
| At3g46060 | RAB-E1c/ARA3 | |
| At3g53610 | RAB-E1a | |
| At5g03520 | RAB-E1d | |
| At3g53710 | AGD6 | ARF GTPase-activating protein AGD6 |
| At4g12120 | SEC1B | Member of KEULE gene family |
| At4g32285 | ENTH/ANTH/VHS superfamily protein | |
| At4g35730 | IST1-LIKE 3 | Regulator of Vps4 activity in the MVB pathway protein |
| At5g52580 | RAB GTPase activator activity | |
| At5g54440 | ATTRS130 | TRAPII tethering factor, CLUB |
-
Table 1—source data 1
Complete list of 14-3-3 epsilon interactors based on two biological replicates.
Proteins listed in Table 1 are shown in bold face while well characterized 14-3-3 clients are highlighted in green.
- https://doi.org/10.7554/eLife.24336.019





