Functionally diverse human T cells recognize non-microbial antigens presented by MR1
Figures
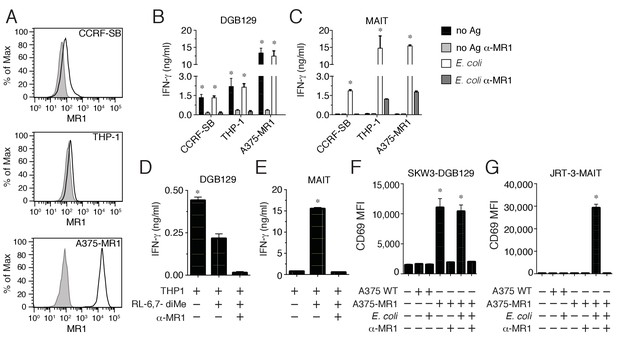
Recognition of non-microbial antigens by MR1-restricted T cells.
(A) Surface expression of MR1 by CCRFSB, THP-1 and A375-MR1 cells. Grey histograms indicate staining with isotype-matched control mAbs. Stimulation of (B) T cell clone DGB129 or (C) MAIT cell clone SMC3 by the three cell lines in A in the absence (no Ag) or presence of E. coli lysate (E. coli) and/or anti-MR1 blocking mAbs (α-MR1). Columns indicate IFN-γ release (mean + SD). Stimulation of (D) DGB129 MR1T or (E) SMC3 MAIT cells by THP-1 cells, constitutively expressing surface MR1, loaded with synthetic 6,7-dimethyl-8-D-ribityllumazine (RL-6,7-diMe) with or without anti-MR1 mAbs. Columns indicate mean IFN-γ release + SD. Stimulation of (F) SKW-3 cells expressing the DGB129 TCR (SKW3-DGB129) or (G) J.RT3-T3.5 cells expressing the MAIT MRC25 clone TCR (J.RT3-MAIT) with A375 cells that expressed (A375-MR1) or lacked (A375-WT) MR1, with or without E. coli lysate and/or anti-MR1 mAbs. CD69 median fluorescence intensity (MFI) ± SD of duplicate cultures of transduced T cells are shown. The CD69 MFI of transduced T cells cultured in the absence of APCs is also shown. Data are representative of four (A, B and C), two (D and E), and three (F and G) independent experiments. *p<0.05 (Unpaired Student’s t-test).
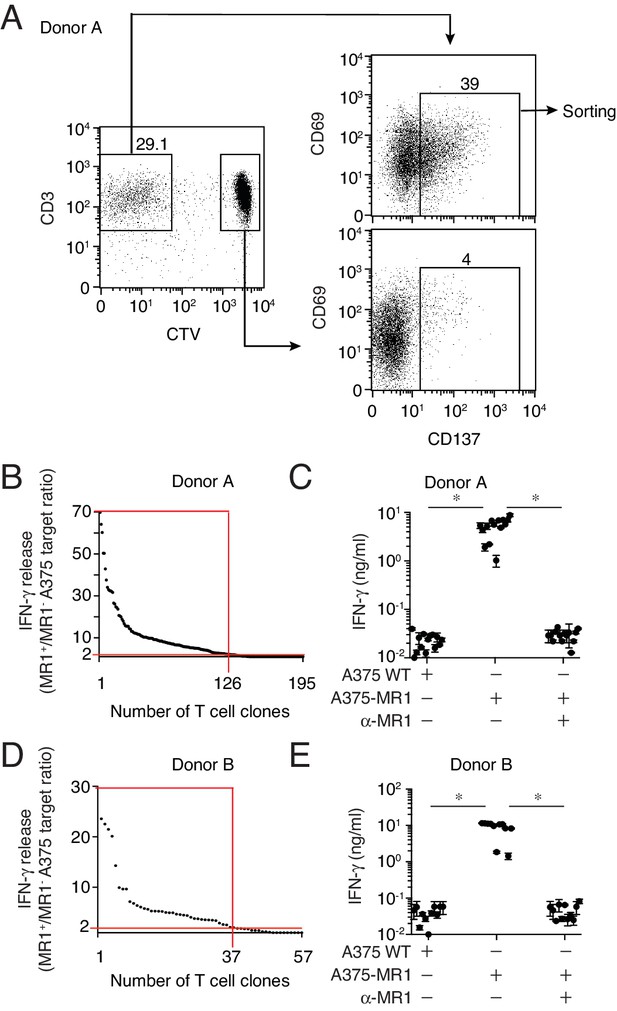
Isolation of non-MAIT MR1-restricted T cell clones after stimulation with A375-MR1 cells in the absence of microbial antigens.
(A) FACS analysis of purified T cells previously expanded with irradiated A375-MR1 cells following overnight co-culture with A375-MR1 cells. Left dot plot shows CD3 and CellTrace violet (CTV) staining in live cells. Upper right and bottom right dot plots show CD69 and CD137 expression on CD3+CTV− and CD3+CTV+ gated cells, respectively. Arrows indicate gating hierarchy. Numbers indicate the percentages of cells within the gates. Cells from Donor A are illustrated as a representative donor. (B, D) Cumulative results of T cell clones screening from Donors A and B. T cell clones were generated from CD3+CTV-CD137high sorted T cells as depicted in A. Graphs show the individual clones (x axis) and their IFN-γ release (y axis), expressed as ratio between the amount of cytokine secreted in response to A375-MR1 cells vs. A375 WT cells. Each dot represents a single T cell clone, tested at the same time in the indicated experimental conditions. The horizontal red line marks the arbitrary IFN-γ ratio cut-off of two, above which MR1-dependent T cell clone reactivity was set. The intercept of the vertical red line indicates the number of MR1-restricted T cell clones in each donor. Red boxes highlight T cell clones whose reactivity was MR1-dependent. Results are representative of two independent experiments. (C, E) IFN-γ release by 14 representative clones from Donor A and 11 clones from Donor B after stimulation with A375 WT, A375-MR1 and A375-MR1 in the presence of blocking anti-MR1 mAbs (α-MR1). Dots represent the IFN-γ release (mean ± SD of duplicate cultures) by each clone. Results are representative of three independent experiments. *p<0.05 (Unpaired Student’s t-test).
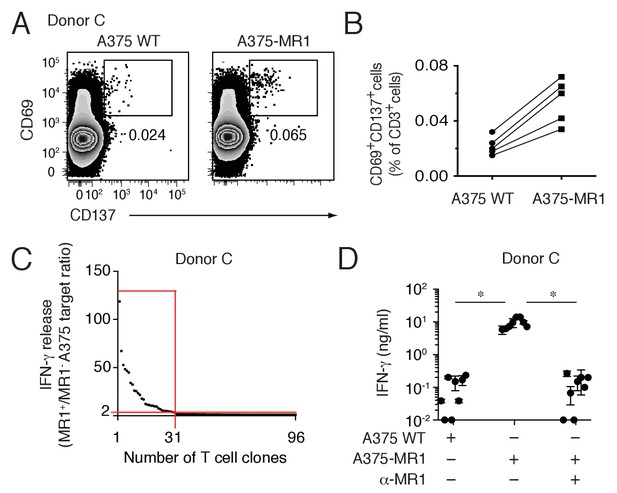
Non-MAIT MR1-restricted T cells are readily detectable in the blood of healthy individuals.
(A) Flow cytometry analysis of purified T cells from a representative donor (Donor C) after overnight co-culture with A375 WT or A375-MR1 cells. Dot plots show CD69 and CD137 expression on live CD3+ cells. Numbers indicate the percentage of cells in the gates. (B) Frequency of CD69+CD137+ T cells from five different donors after overnight co-culture with A375 WT or A375-MR1 cells. (C) Cumulative results of T cell clone stimulation assays from Donor C. T cell clones were generated from CD3+CD69+CD137+ sorted T cells as depicted in A, right dot plot. The graph shows the number of tested clones (x axis) and IFN-γ release (y axis) expressed as ratio between the amount of cytokine secreted in response to A375-MR1 cells vs. A375-WT cells. Each dot represents a single T cell clone, tested at the same time in the indicated experimental conditions. The horizontal red line marks the arbitrary IFN-γ ratio threshold of two above which MR1-dependent T cell clone reactivity was set. The intercept of the vertical red line indicates the number of MR1-restricted T cell clones. Red box highlights T cell clones whose reactivity was MR1-dependent. Results are representative of two independent experiments. (D) Recognition of A375-MR1 but not A375 WT cells in the absence of exogenous antigens by eight representative MR1-restricted T cell clones from Donor C. Inhibition of T cell clone reactivity to A375-MR1 cells by blocking anti-MR1 mAbs (α-MR1). Dots represent the IFN-γ release (mean ± SD of duplicate cultures) by each clone tested in the three experimental conditions. Results are representative of three independent experiments. *p<0.05 (Unpaired Student’s t-test).
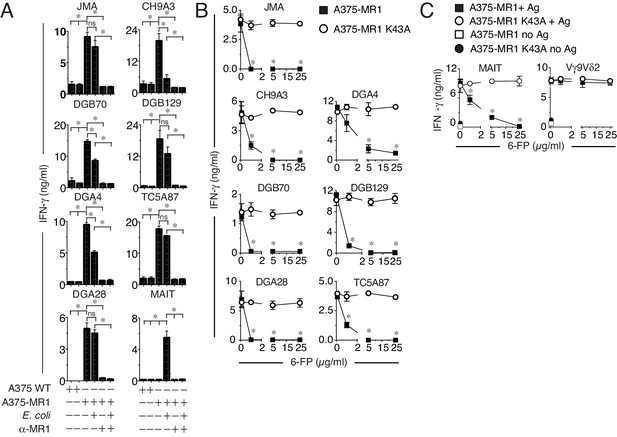
MR1T cell clones do not react to microbial ligands or to 6-FP.
(A) Response of seven MR1T cell clones and one control MAIT cell clone co-cultured with A375 cells expressing (A375-MR1) or not (A375 WT) MR1 in the presence or absence of E. coli lysate. Blocking of T cell clone reactivity by anti-MR1 mAbs (α-MR1) is also shown. (B) Response of MR1T cell clones to A375 cells expressing either WT MR1 molecules (A375-MR1) or K43A-mutated MR1 molecules (A375-MR1 K43A) in the presence of 6-formyl pterin (6-FP). (C) Stimulation of control MAIT cell clone MRC25 or control TCR Vγ9Vδ2 clone G2B9 with A375-MR1 or A375-MR1 K43A cells previously incubated with or without E. coli lysate or zoledronate, respectively, either in the absence or presence of 6-FP. Results are expressed as mean ± SD of IFN-γ measured in duplicate cultures. Results are representative of three independent experiments. *p<0.05 (Unpaired Student’s t-test).
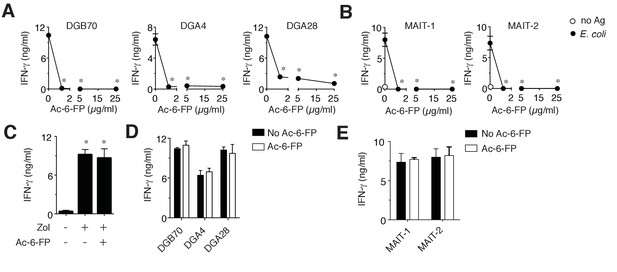
MR1T cell clones do not recognize Ac-6-FP.
(A) Stimulation of three representative MR1T cell clones by A375-MR1 cells in the absence or presence of acetyl-6-formyl pterin (Ac-6-FP). (B) Stimulation of two MAIT cell clones (MRC25 and SMC3) by A375-MR1 cells pulsed with E. coli lysate in the absence or presence of Ac-6-FP. (C) A375-MR1 cells were treated with zoledronate (Zol) in the absence or presence of Ac-6-FP (25 µg/ml) and used to stimulate a TCR Vγ9-Vδ2 cell clone (G2B9). (D) A375 cells expressing K43A mutant MR1 molecules (A375-MR1 K43A) were used to stimulate the three MR1T cell clones shown in A, in the absence or presence of Ac-6-FP (25 µg/ml). (E) Stimulation of the two MAIT cell clones used in B by A375-MR1 K43A cells pulsed with E. coli lysate in the absence or presence of Ac-6-FP (25 µg/ml). Results are expressed as mean ± SD of IFN-γ release assessed in duplicate cultures and are representative of three independent experiments. *p<0.05 (Unpaired Student’s t-test).
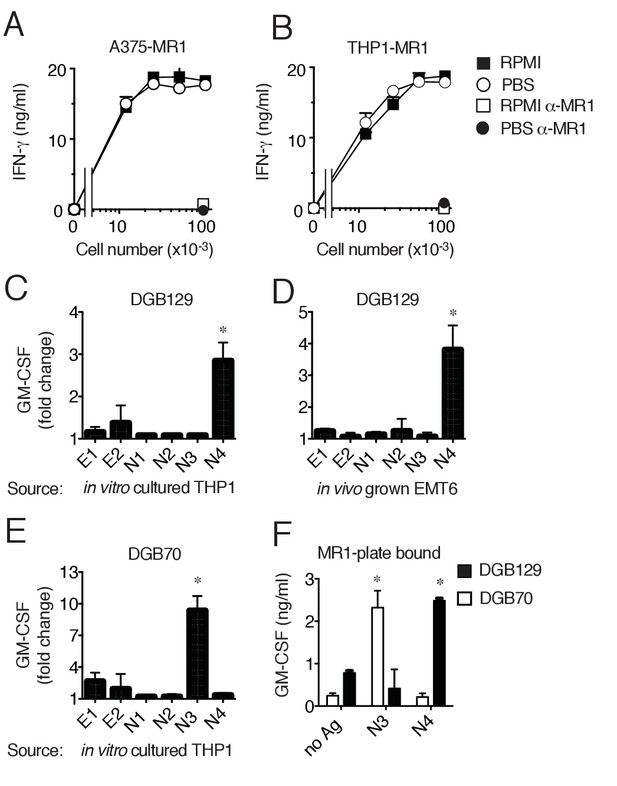
MR1T cells recognize diverse antigens not derived from RPMI 1640 medium.
Stimulation of the DGB129 MR1T cell clone by MR1-overexpressing (A) A375 cells (A375-MR1) and (B) THP-1 cells (THP1-MR1) grown for 4 days in RPMI 1640 or in PBS both supplemented with 5% human serum. Inhibition of T cell clone reactivity by anti-MR1 blocking mAbs (α-MR1) is shown. DGB129 cells recognize APCs loaded with fractions isolated from (C) THP-1 cell lysate or from (D) in vivo grown mouse breast tumor EMT6. Fractions E1 and E2 contain hydrophobic molecules; fractions N1-N4 contain hydrophilic molecules. (E) DGB70 MR1T cells react to N3 fraction of THP-1 lysate. (F) Stimulation of DGB129 and DGB70 T cells by THP-1-derived fractions N3 and N4 loaded onto plastic-bound recombinant MR1. Shown is T cell release of IFN-γ or GM-CSF mean ± SD of duplicate cultures (representative of three independent experiments). Total cytokine release is shown in panels A, B, F; fold increase over background is shown in panels C, D, E. *p<0.05 (Unpaired Student’s t-test).
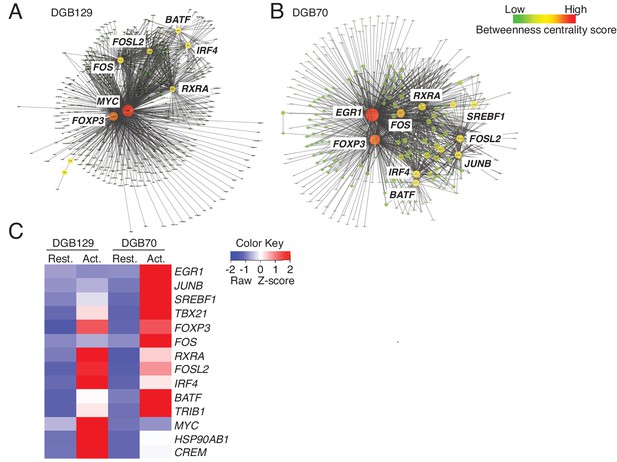
MR1T cell clones exhibit divergent transcriptional responses to antigen stimulation.
Betweeness centrality analysis illustrating the key transcription factors that were differentially expressed in resting (No Ag) and antigen-activated (Ag Act) MR1T cell clones (A) DGB129 and (B) DGB70. Color code represents betweeness centrality score. The size of the nodes (or hubs) indicates the relative importance of individual transcription factors within the whole gene network. (C) Heat-map comparing key transcription factors that were differentially expressed in resting (Rest.) and antigen-activated (Act.) DGB129 and DGB70 MR1T cell clones (analysis performed by PageRank algorithm).
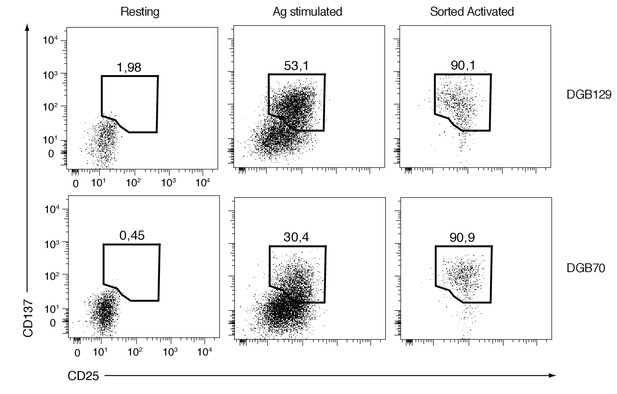
FACS analysis of resting and activated DGB129 and DGB70 MR1T cells used for transcriptome studies.
Dot-plots display the expression of CD25 and CD137 activation markers on T cells cultured alone (Resting) or stimulated with A375-MR1 cells (Ag stimulated). Purity of CD25+CD137+ cells sorted from the Ag stimulated groups (Sorted Activated) is shown. Plots are gated on CD3+ DAPI- cells. Resting and Sorted Activated DGB129 and DGB70 MR1T cells were utilized for next-generation sequencing experiments.
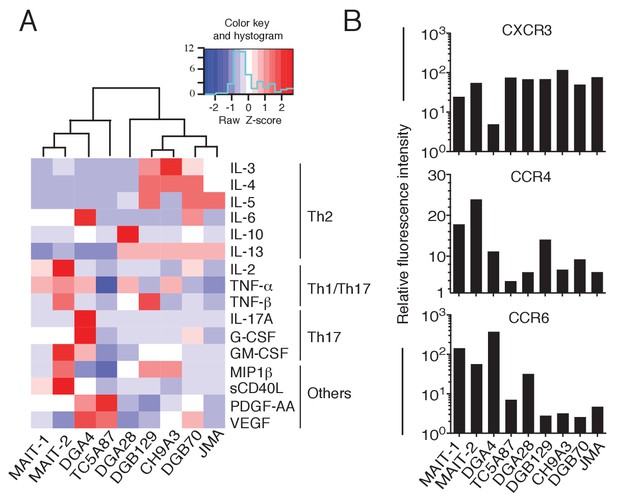
Functional heterogeneity of MR1T cell clones.
(A) Heat-map of cytokine expression by seven different MR1T cell clones when stimulated by MR1-expressing A375 cells. Also shown are the cytokine profiles of control MAIT cell clones MRC25 (MAIT-1) and SMC3 (MAIT-2) following activation by A375-MR1 cells pulsed with E. coli lysate. Cytokines were assessed in the supernatants of duplicate cultures. The mean value for each cytokine was used to generate the heat-map. Cluster analysis was performed using Pearson correlation. Graphs displaying the amounts of individual cytokines released by the different clones are shown in Figure 7—figure supplement 1. (B) Flow cytometry analysis of CXCR3, CCR4 and CCR6 surface expression by resting MR1T cell clones and 2 MAIT control clones (MRC25, MAIT-1 and SMC3, MAIT-2). Graphs show the relative fluorescence intensity calculated by dividing the median fluorescence intensity (MFI) of specific mAb staining by the MFI of the corresponding isotype control. Data are representative of two independent experiments.
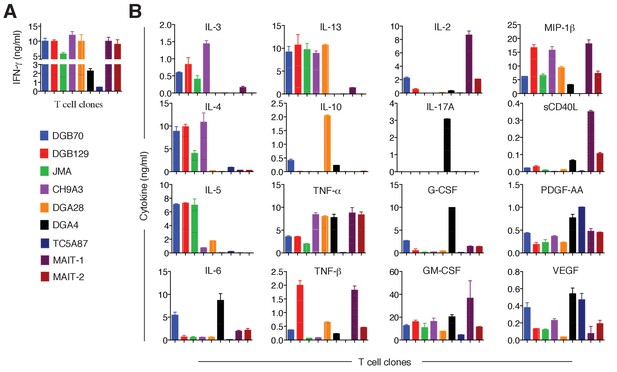
Cytokines released by antigen-stimulated MR1T and MAIT cell clones.
(A) IFN-γ released by 7 MR1T cell clones stimulated with A375-MR1 cells and 2 MAIT cell clones (MRC25 and SMC3) stimulated with A375-MR1 cells pulsed with E. coli lysate. ELISA results are expressed as mean ± SD of IFN-γ release measured in duplicate cultures. (B) Analysis of 16 additional cytokines by multiplex cytokine assay performed on the same supernatants for which IFN-γ is shown in A. Results are representative of two independent experiments.
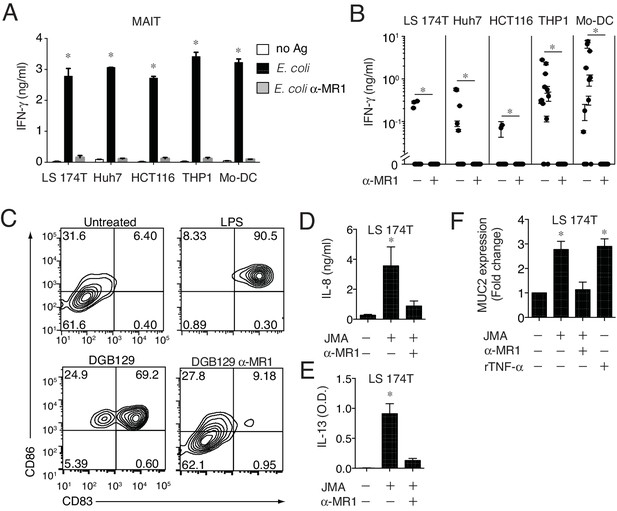
MR1T cells recognize cells constitutively expressing low surface MR1 and show diverse T helper-like functions.
(A) Recognition of four human cells lines expressing constitutive surface levels of MR1 and Mo-DCs by the representative SMC3 MAIT cell clone in the absence (no Ag) or presence of E. coli lysate (E. coli) with or without anti-MR1 blocking mAbs (α-MR1). (B) Recognition of the same cell types as in A by thirteen MR1T cell clones with or without anti-MR1 mAbs (α-MR1). Graphs show IFN-γ release (mean ±SD of duplicate cultures). (C) Flow cytometry analysis of co-stimulatory molecules CD83 and CD86 on Mo-DCs after co-culture with DGB129 MR1T cells with or without anti-MR1 mAbs (α-MR1). A control group consisting of Mo-DCs stimulated with LPS (10 ng/ml) in the absence of T cells is also shown. Numbers indicate percentages of cells in each quadrant. (D,E) Stimulation of JMA MR1T cells by LS 174T intestinal epithelial cells with or without anti-MR1 mAbs (α-MR1). Columns show (D) IL-8 (ng/ml) and (E) IL-13 (optical density, O.D.) release. (F) Q-PCR analysis of MUC2 gene expression in LS 174 T cells cultured alone or with JMA MR1T cells in the presence or absence of anti-MR1 mAbs (α-MR1). As control, LS 174 T cells were stimulated with recombinant TNF-α (rTNF-α, 10 ng/ml) in the absence of MR1T cells. All data are expressed as mean ± SD of triplicate cultures. Results are representative of at least three independent experiments. *p<0.05 (Unpaired Student’s t-test).
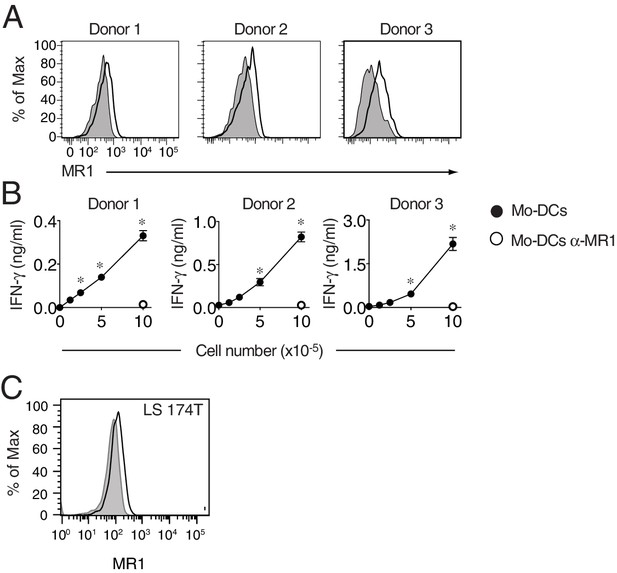
MR1 surface expression by monocyte-derived dendritic cells and LS 174 T cells.
(A) Flow cytometry detection of MR1 surface protein expression on monocyte-derived dendritic cells (Mo-DCs) generated from blood of three healthy donors. Grey histograms represent staining with isotype-matched control mAbs. (B) Recognition of Mo-DCs from the three donors in A by DGB129 MR1T cell clones in the absence or presence of anti-MR1 (α-MR1) mAbs. IFN-γ release in the supernatants is shown and expressed as mean ± SD. Results are representative of three independent experiments. *p<0.05 (Unpaired Student’s t-test). (C) MR1 surface protein expression by LS 174T intestinal epithelial cells. Grey histograms represent staining with isotype-matched control mAbs.
Tables
Phenotype and TCR gene usage of selected MR1-reactive T cell clones.
| Clone | CD4 | CD8α | TCRα | TCRβ | CD161 |
|---|---|---|---|---|---|
| DGB129 | − | + | TRAV29 | TRBV12-4 | − |
| DGB70 | − | − | TRAV5 | TRBV28 | − |
| DGA28 | − | + | TRAV25 | TRBV29-1 | + |
| DGA4 | − | − | TRAV1-2 | ND | + |
| JMA | − | + | TRAV27 | TRBV25-1 | − |
| TC5A87 | − | + | TRAV13-1 | TRBV25-1 | − |
| CH9A3 | − | + | TRAV24 | TRBV5-5 | − |
-
ND, not determined.
Additional files
-
Supplementary file 1
Genes modulated in activated vs. resting MR1T cell clones.
- https://doi.org/10.7554/eLife.24476.016





