ERG-28 controls BK channel trafficking in the ER to regulate synaptic function and alcohol response in C. elegans
Figures
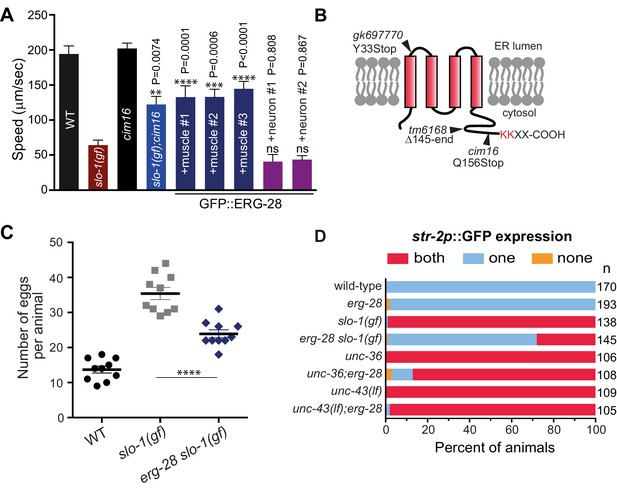
Loss-of-function mutation in erg-28 suppresses phenotypic defects of slo-1 gain-of-function mutant.
(A) Mutation in erg-28 suppresses sluggish locomotion of slo-1(ky399gf) mutant. Average speed of animals with indicated genotypes on NGM agar plates without food. slo-1(gf) and erg-28 alleles are ky399 and cim16, respectively. +muscle #1, #2, #3: three independent lines of erg-28 slo-1(gf);cimEx[myo-3p::gfp::erg-28, ttx-3p::mRFP]. +neuron#1, #2: two independent transgenic lines of erg-28 slo-1(gf);cimEx[H20p::gfp::erg-28, ttx-3p::mRFP]. Error bars represent S.E.M. (standard error of the mean). n = 10. One way ANOVA with Tukey’s post-hoc analysis. P values are presented in comparison to slo-1(gf). (B) ERG-28 is predicted to be an endoplasmic reticulum resident protein with four transmembrane domains and KKXX ER retrieval COPI binding motif. Mutation sites of cim16, tm6168, and gk697770 alleles are indicated. (C) Suppression of egg laying defects of slo-1(ky399gf) mutant by erg-28(cim16) mutation. Number of eggs retained in the animals. Error bars represent S.E.M. n = 10. ****p<0.0001, one way ANOVA with Tukey’s post-hoc analysis. (D) Suppression of defective asymmetric str-2 gene expression in slo-1(ky399gf) mutant by erg-28(gk697770). Mutants used in this experiment: erg-28(gk697770), slo-1(ky399), unc-36(e251), and unc-43(n498n1186). (Figure 1—source data 1: This file contains raw measurement data and statistical analysis summary for Figure 1A, C and D).
-
Figure 1—source data 1
erg-28 mutation suppresses phenotipic defects of slo-1(gf) mutant.
Raw data and statistics for the speed measurement (A) and egg laying assay (C).
- https://doi.org/10.7554/eLife.24733.003
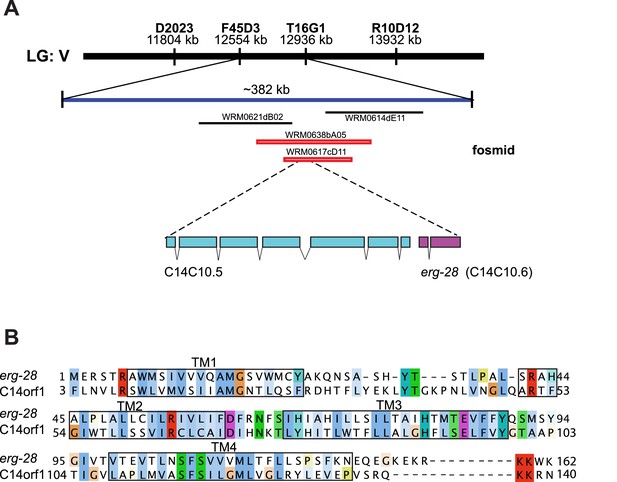
Identification of cim16 mutation in erg-28 gene.
(A) Genetic mapping and fosmid rescue were used to identify the genetic lesion of cim16. Genetic mapping was performed using single nucleotide polymorphisms (SNPs) present between Bristol N2 and Hawaiian CB4856 strains. The erg-28 gene is a part of an operon with an upstream gene C14C10.5. (B) Alignment of the predicted erg-28 coding sequence with the human homolog C14orf1. Alignment was performed using Jalview with Clustal X color scheme (http://www.jalview.org/help/html/colourSchemes/clustal.html) (Waterhouse et al., 2009). The predicted transmembrane domains (TM1~TM4) are boxed. The predicted ER retention/retrieval sequence motif (KKXX-COOH) is conserved in C. elegans and humans.
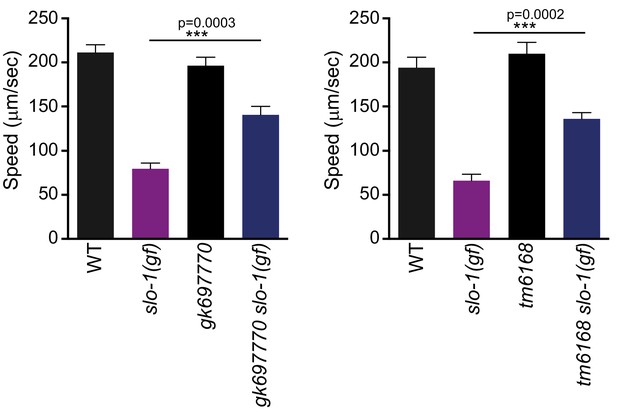
Both gk697770 and tm6168 alleles of erg-28 suppress the sluggish movement of slo-1(ky399) gain-of-function mutant.
Speed measurement was performed as in Figure 1A. Error bars represent S.E.M. (standard error of the mean). n = 10. One way ANOVA with Tukey’s post-hoc analysis.
-
Figure 1—figure supplement 2—source data 1
Two other alleles of erg-28 suppress the locomotory defect of slo-1(gf) mutant.
Raw data and statistics for the speed measurement.
- https://doi.org/10.7554/eLife.24733.006
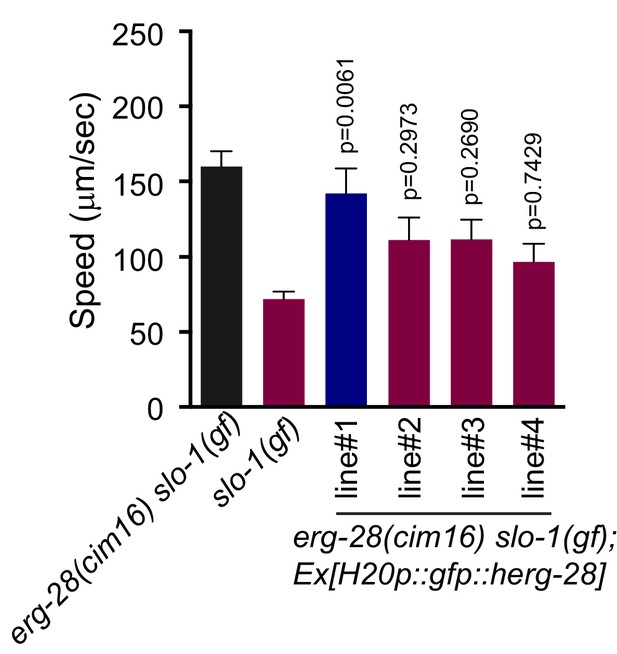
The human homolog C14orf1 (herg-28) can partially replace C. elegans erg-28.
Speed measurement was performed as in Figure 1A. C14orf1(herg-28) was expressed under the control of the pan-neuronal promoter in erg-28(cim16) slo-1(ky399gf) double mutant animals. The speeds of four different lines were measured. Error bars represent S.E.M. (standard error of the mean). n = 10. One way ANOVA with Tukey’s post-hoc analysis. P values are presented in comparison to slo-1(gf).
-
Figure 1—figure supplement 3—source data 1
Functional conservation of human homolog with C. elegans erg-28.
Raw data and statistics for the speed measurement.
- https://doi.org/10.7554/eLife.24733.008
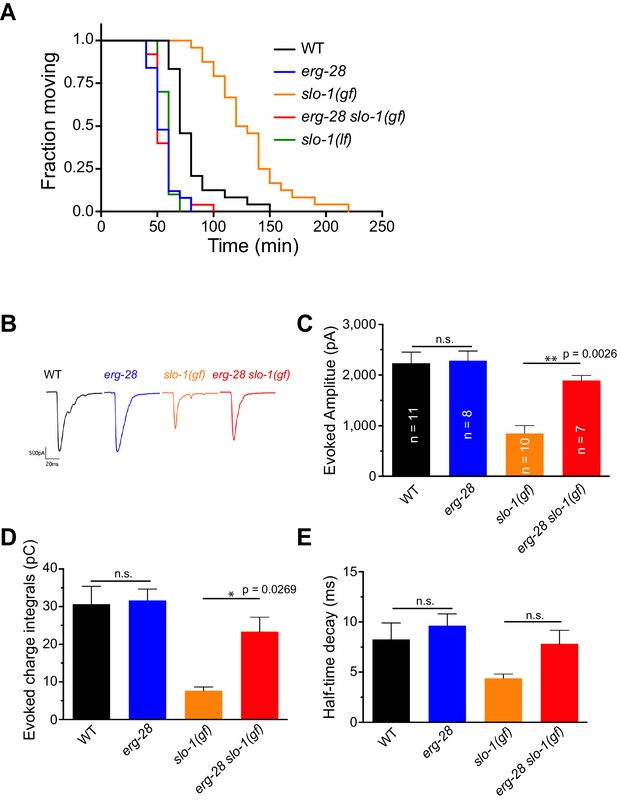
ERG-28 is required for SLO-1 function in synaptic transmission.
(A) erg-28(gk697770) mutation suppresses aldicarb resistance of slo-1(ky399gf) mutant and is aldicarb hypersensitive. Aldicarb-induced paralysis was analyzed using Kaplan-Meier survival analysis. Three independent trials with n > 20 for each genotype in each trial. All three trials showed similar results. Trial 1 is shown and statistics of 3 trials is presented in Figure 2-source data 1. Mutants used are erg-28 (gk697770), slo-1(ky399), and slo-1(eg142 loss of function). 1 mM aldicarb was used. (B) Representative evoked responses from voltage-clamped body wall muscles (holding potential −60 mV) in response to 2 ms blue-light activation of neuronally expressed channelrhodopsin-2. (C–E) erg-28(gk697770) suppresses the synaptic defects of slo-1(ky399gf) mutants. n.s.: not significant, One way ANOVA with Tukey’s post-hoc analysis. Figure 2—source data 1: This file contains raw measurement data and statistical analysis summary for Figure 2C,D and E.
-
Figure 2—source data 1
erg-28 mutation suppresses the synaptic transmission defect of slo-1(gf) mutant.
Raw data and statistics for the aldicarb assay (A) and the electrophysiology (B-E).
- https://doi.org/10.7554/eLife.24733.010
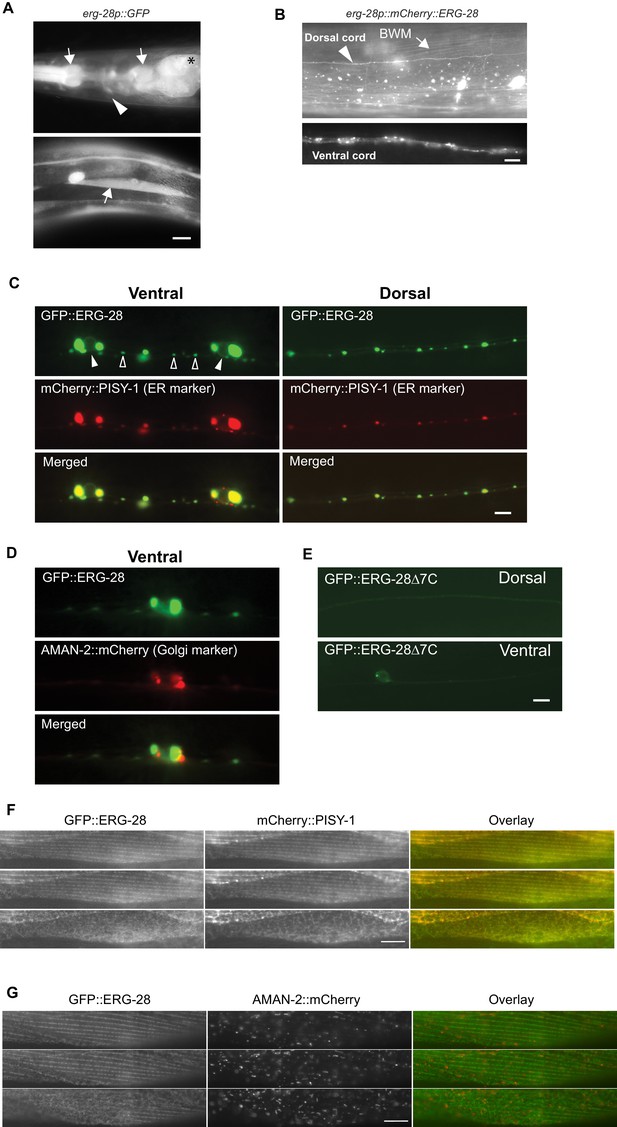
ERG-28 is an endoplasmic reticulum resident protein expressed in muscles and neurons.
(A) erg-28 is a part of an operon with C14C10.5. 2.5 kb upstream of C14C10.5 gene including 30 bp of coding region of C14C10.5 was fused with GFP (cimEx54[erg-28p::gfp, ttx-3p::mRFP]). Expression was observed in muscles (arrows), neurons (arrowheads), and intestines (asterisk). Scale bar: 10 μm. (B) ERG-28 is prominently expressed in muscles (arrow) and neurons (arrowhead). mCherry-tagged ERG-28 (cimIs39[erg-28p::mCherry::erg-28, odr-1p::gfp]) was expressed under the same promoter as in Figure 3A. Scale bar: 10 μm. (C) ERG-28 co-localizes with the ER marker PISY-1 in a subset of motor neurons. GFP::ERG-28 and PISY-1::mCherry were expressed using the unc-129 neuronal promoter (cimEx63[unc-129p::mCherry::pisy-1, unc-129p::gfp::erg-28]). Closed arrowheads: neuron nuclei. Open arrowheads: puncta at dendrites. Scale bar: 10 μm. (D) ERG-28 is minimally co-localized with the Golgi marker AMAN-2 in a subset of motor neurons. GFP::ERG-28 and AMAN-2::mCherry were expressed using the unc-129 neuronal promoter (cimEx59[unc-129p::aman-2::mCherry, unc-129p::gfp::erg-28]). Scale bar: 10 μm. (E) The last 7 amino acids of ERG-28 are required for stable expression of ERG-28. ERG-28::GFP with 7 amino acid deletion was expressed using unc-129 neuronal promoter (cimEx66[unc-129p::gfp::erg-28Δ7, ttx-3p::mRFP]). Scale bar: 10 μm. (F) GFP::ERG-28 co-localizes with the ER marker in muscle cells. GFP::ERG-28 and mCherry::PISY-1 were expressed using the myo-3 promoter (cimEx88[myo-3p::gfp::erg-28, myo-3p::mCherry::pisy-1, rol-6(d)]). Scale bar: 10 μm. Three different sections from the z-stack are shown. (G) GFP::ERG-28 does not co-localize with the Golgi marker in muscle cells. GFP::ERG-28 and AMAN-2::mCherry were expressed using the myo-3 promoter (cimEx91[myo-3p::gfp::erg-28, myo-3p::aman-2::mCherry, rol-6(d)]). Scale bar: 10 μm. Three different sections from the z-stack are shown. Images of additional sections are shown in Figure 3—figure supplement 1.
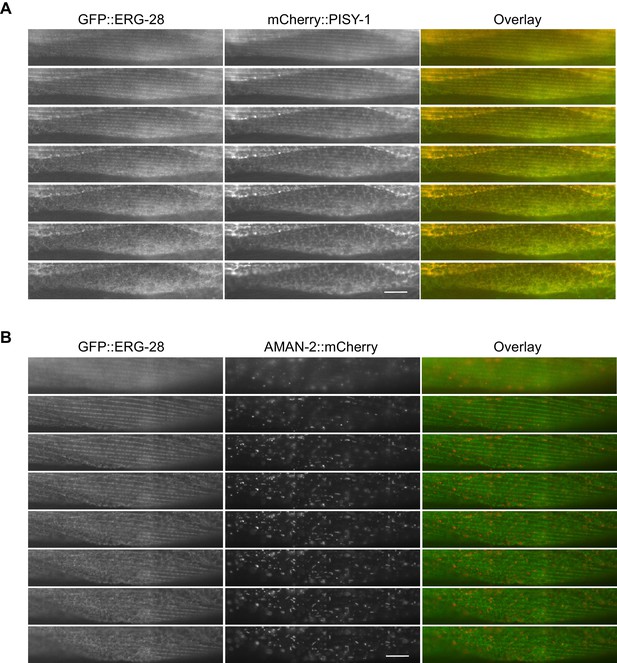
GFP::ERG-28 co-localizes with the ER marker (A), but not with the Golgi marker (B) in muscle cells.
Muscle cells with a low expression level of GFP::ERG-28 were imaged using the z-scanning acquisition (0.2 micron intervals) from the muscle plasma membrane (top) to the muscle interior/muscle belly (bottom). (A) cimEx88[myo-3p::gfp::erg-28, myo-3p::mCherry::pisy-1, rol-6(d)]. (B) cimEx91[myo-3p::gfp::erg-28, myo-3p::aman-2::mCherry, rol-6(d)].
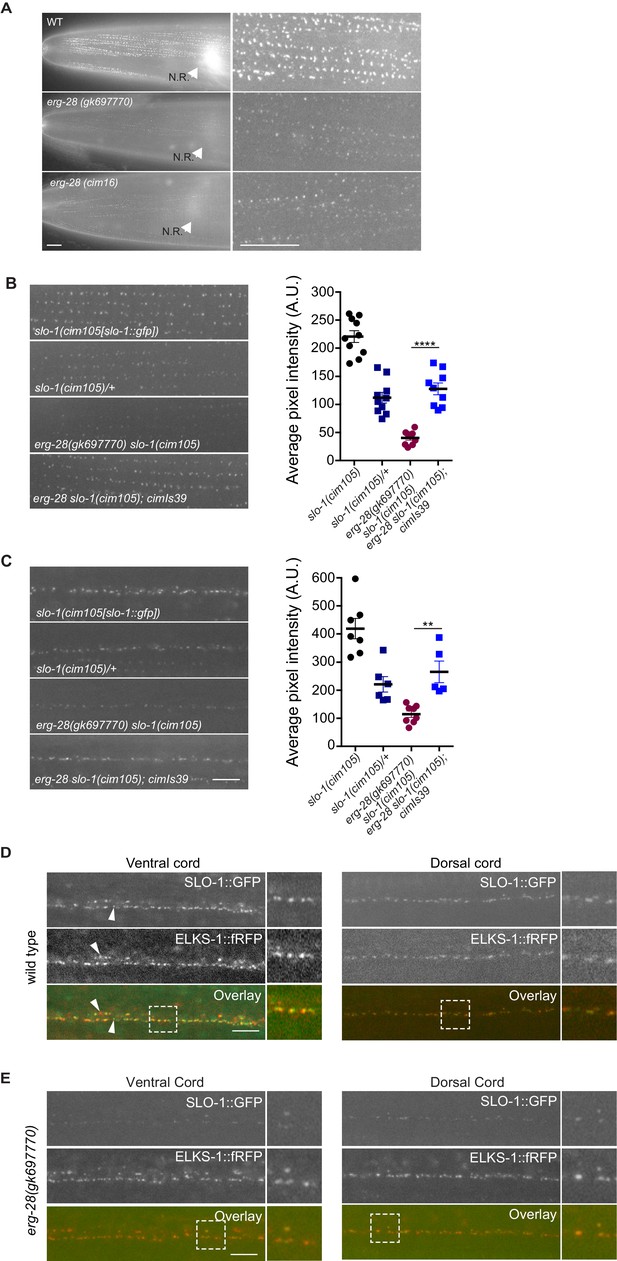
ERG-28 is required for the normal expression level of SLO-1 at synaptic terminals and muscle excitation sites.
(A) SLO-1::GFP expression is reduced in erg-28(gk697770) and erg-28(cim16) mutants. N.R: nerve ring. Scale bar: 10 μm. (B and C) SLO-1 expression is restored by expression of ERG-28 in muscle cells (B) and neurons (C). Right panels are quantification of puncta in muscles (B) and the dorsal cords (C). slo-1(cim105)/+ heterozygote animals were generated by crossing slo-1(cim105) with wild-type males. cimIs39[erg-28p::mCherry::erg-28, odr-1p::gfp] is the chromosomally integrated array expressing mCherry::ERG-28 from the erg-28 promoter. (D) SLO-1::GFP is expressed at presynaptic terminals and co-localizes with active zone marker, ELKS-1::fRFP in the nerve cords. Occasionally, puncta that do not co-localize with each other are observed (arrowheads). Dash lined boxes indicate the areas that are zoomed in and shown in the right. Scale bar: 10 μm. (E) In erg-28(gk697770) mutant, SLO-1::GFP expression is drastically reduced, but the remaining SLO-1::GFP co-localizes with ELKS-1::fRFP in the nerve cords. Scale bar: 10 μm.
-
Figure 4—source data 1
SLO-1 expression in the muscle (B) and neuron (C) is reduced in erg-28 mutant.
Raw data and statistics for the fluorescence measurement.
- https://doi.org/10.7554/eLife.24733.015
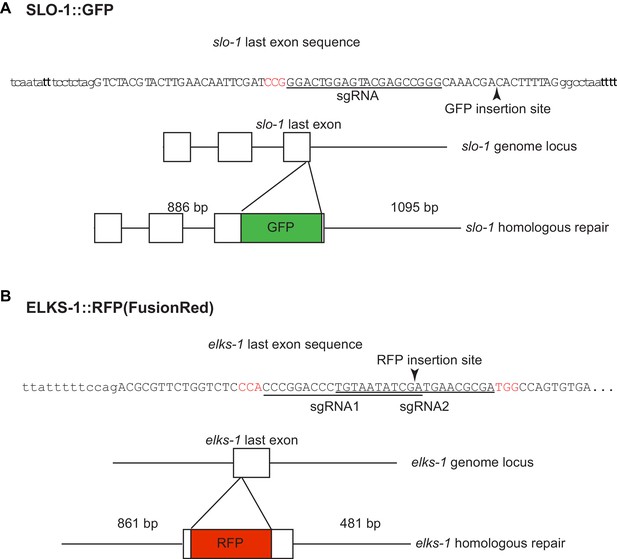
Introduction of GFP and fRFP (FusionRed) into the slo-1 (A) and elks-1 (B) genomic loci using CRISPR/Cas9 technology.
sgRNA sequences are underlined and PAM sequences are shown in red.
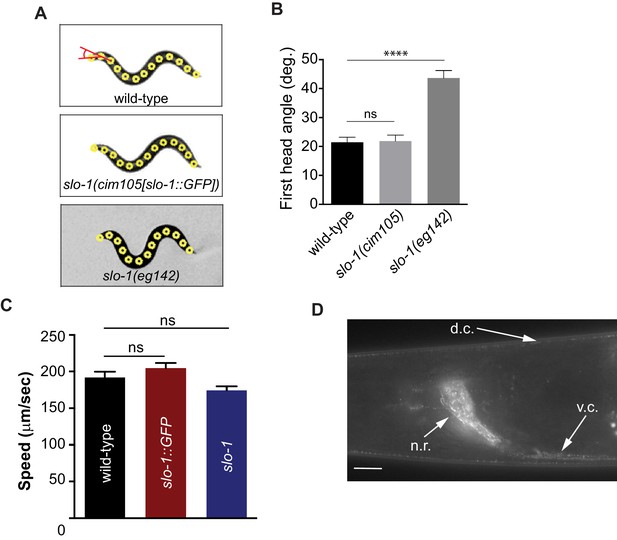
Introduction of GFP at the end of the genomic slo-1 coding sequence does not interfere with head bending angle (A and B) or normal locomotory speed (C).
The data are presented as the mean ± S.E.M. ns, not significant (p>0.05); one-way ANOVA with Dunnett’s post hoc analysis. (D) SLO-1::GFP is expressed as distinct puncta in neurons and muscles. d.c. dorsal cord, v.c., ventral cord, n.r, nerve ring. Scale bar: 10 μm. Figure 4—figure supplement 2—source data 1: This file contains raw measurement data and statistical analysis summary for Figure 4—figure supplement 2.
-
Figure 4—figure supplement 2—source data 1
The C-terminal GFP fusion does not interfere with normal function of SLO-1.
Raw data and statistics for the head angle (A) and the speed (C) measurement.
- https://doi.org/10.7554/eLife.24733.018
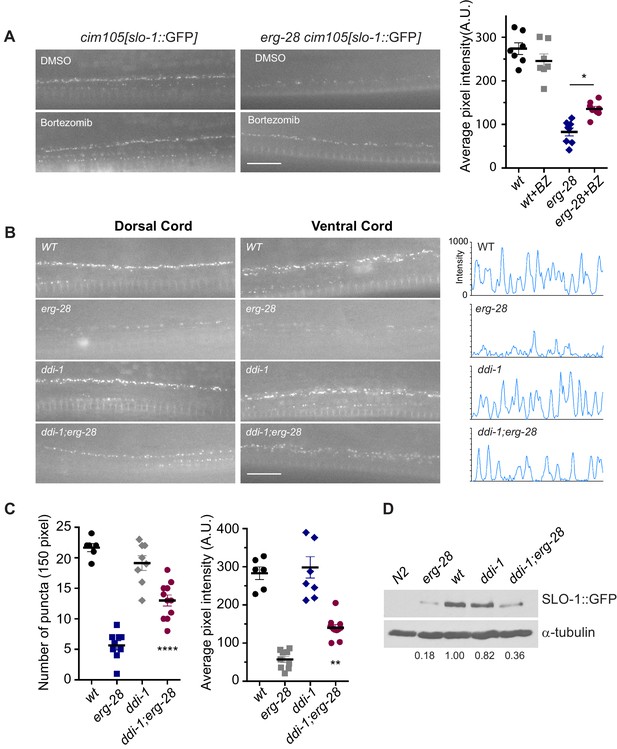
SLO-1 is degraded by proteasome in the absence of ERG-28.
(A) Proteasome inhibition with bortezomib (BZ) partially restores SLO-1 expression at the presynaptic terminals in erg-28(gk69777) mutant. Scale bar: 10 μm. Right panel: SLO-1 average pixel intensity in the dorsal cord of DMSO or BZ treated strains was quantified. Error bars represent S.E.M. *p=0.0112, one way ANOVA with Tukey’s post-hoc analysis. (B) ddi-1 mutation partially restores SLO-1 expression in erg-28 mutant. Right panel: Intensity profiles of the dorsal cords (150 pixels) in wild-type, erg-28(gk697770), ddi-1(ok1468) and ddi-1(ok1468);erg-28(gk697770) mutant animals. Average pixel intensity of SLO-1 in the dorsal cord was quantified. (C) The number and intensity of SLO-1 puncta in wild-type, erg-28(gk697770), ddi-1(ok1468) and ddi-1(ok1468);erg-28(gk697770) mutant animals were quantified. Error bars represent S.E.M. **p=0.0021 erg-28 vs. ddi-1;erg-28, ****p<0.0001, erg-28 vs. ddi-1;erg-28, one way ANOVA with Tukey’s post-hoc analysis. (D) Western analysis shows that total SLO-1::GFP amount is reduced in erg-28(gk697770) mutant and partially restored in ddi-1(ok1468);erg-28(gk697770) double mutant. SLO-1::GFP intensity was normalized to the α-tubulin and the relative intensities compared to the wild type were indicated. N2 does not express GFP and is a negative control for western blotting with anti-GFP antibody. Figure 5—source data 1: This file contains raw measurement data and statistical analysis summary for Figure 5.
-
Figure 5—source data 1
Proteasome inhibition and ddi-1 mutation partially restore SLO-1 expression in erg-28 mutant.
Raw data and statistics for the fluorescence measurement.
- https://doi.org/10.7554/eLife.24733.020
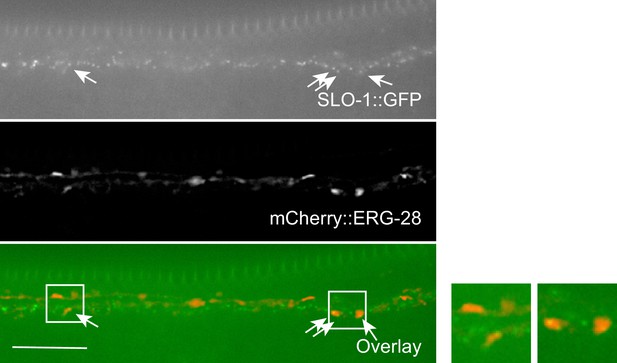
Co-localization of SLO-1 and ERG-28 in the ventral cord.
ERG-28 and SLO-1 occasionally co-localize together. Scale bar: 10 μm.
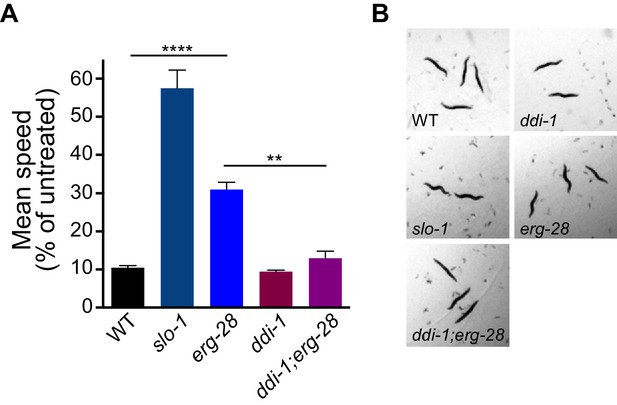
erg-28 mutation confers the resistance to the intoxicating effect of ethanol and ddi-1 mutation reverses the ethanol-resistant behavior.
(A) The locomotory speed was measured in the presence of ethanol and its values are divided by the average speed of untreated animals. Error bars represent S.E.M. **p=0.0033, ****p<0.0001, one way ANOVA with Tukey’s post-hoc analysis. See Videos 2–4. (B) The snapshots of animals in the presence of intoxicating animals show sinusoidal posture in slo-1(eg142lf), erg-28(gk697770), ddi-1(ok1468), and ddi-1(ok1468);erg-28(gk697770) mutants. Figure 6—source data 1: This file contains raw measurement data and statistical analysis summary for Figure 6.
-
Figure 6—source data 1
ddi-1 mutation reverses alcohol-resistant locomotion of erg-28 mutant.
Raw data and statistics for the speed measurement.
- https://doi.org/10.7554/eLife.24733.023
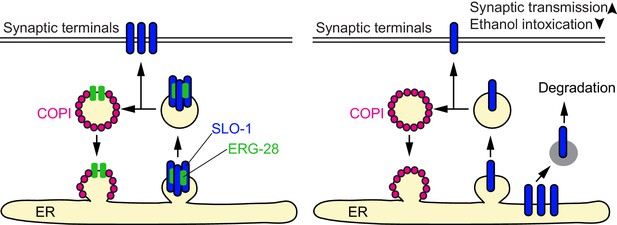
A model for ERG-28 function.
ERG-28 as a chaperone for the SLO-1 channel complex in the ER. In the absence of ERG-28, SLO-1 is not efficiently delivered to the synaptic terminals, and is highly susceptible to DDI-1 and proteasome-dependent degradation.
Videos
ERG-28 is mobile.
mCherry::ERG-28 was expressed using erg-28 promoter, cimIs39[erg-28p::mCherry::erg-28, odr-1p::gfp]. Time lapse image. A movie recorded for 14 s.
A movie for wild-type, erg-28(gk697770) and slo-1(eg142) in the presence of intoxicating dose of ethanol.
https://doi.org/10.7554/eLife.24733.025A movie for wild-type, erg-28(gk697770) and ddi-1(ok1468);erg-28(gk697770) in the presence of intoxicating dose of ethanol.
https://doi.org/10.7554/eLife.24733.026A movie for wild-type, slo-1(eg142) and ddi-1(ok1468) in the presence of intoxicating dose of ethanol.
https://doi.org/10.7554/eLife.24733.027Additional files
-
Supplementary file 1
Lists of plasmid constructs and strains used in this study.
- https://doi.org/10.7554/eLife.24733.028





