LARP1 functions as a molecular switch for mTORC1-mediated translation of an essential class of mRNAs
Figures
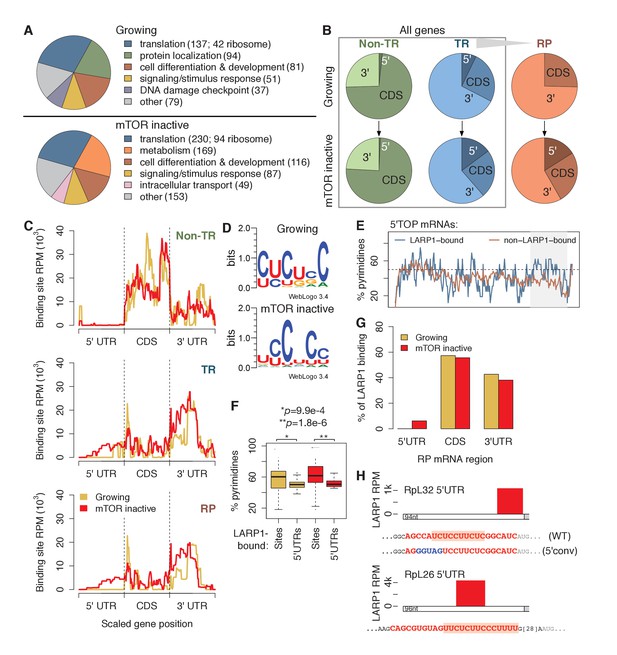
LARP1 binds pyrimidine-rich 5’UTR regions of translation-related transcripts.
(A) LARP1-bound genes are most enriched for GO terms related to translation including RP genes. (B) Upon mTOR inactivation, LARP1 binding at 5’UTRs increases on TR and RP genes. (C) LARP1 binding at TR and RP 5’UTRs under mTOR-inactive conditions tends to occur at the 3’ end. (D) LARP1 binds directly to pyrimidine-enriched sequences in 5’UTRs. (E) The LARP1 binding sites at the 3’ end of 5’TOP-containing 5’UTRs are enriched for pyrimidines. (F) LARP1-bound sites on 5’UTRs are enriched for pyrimidines compared to the rest of the 5’UTR sequence. Welch’s two-tailed t-test: *p=1.4e-15 and **p=1.2e-18. (G) LARP1 binding on RP-encoding mRNAs is gained at 5’UTRs upon mTOR inactivation and slightly decreased at CDSs and 3’UTRs. (H) The locations (red box), sequences (red color), and motifs (orange background) of the 5’ UTR LARP1 binding site in RpL32 and RpL26 mRNAs under mTOR-inactive conditions. Substituted nucleotides are highlighted by blue color.
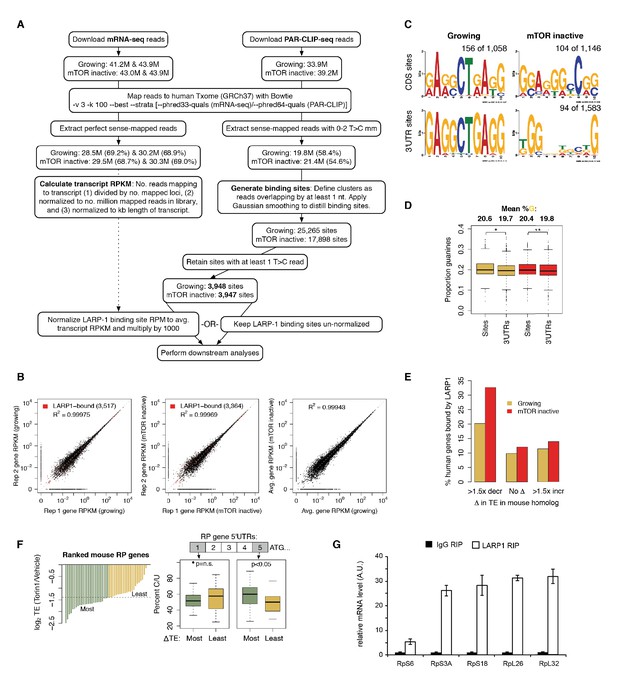
LARP1 binds 5’UTR pyrimidine-rich regions of translation-related transcripts.
(A) Schematic of computational steps used to derived LARP1 binding sites from PAR-CLIP- seq and mRNA-seq libraries. (B) Reproducibility of replicate mRNA-seq libraries. Gene RPKM values are highly correlated between replicate mRNA-seq libraries generated under growing and mTOR inactive conditions. Pearson correlation coefficients are shown on the graphs. Genes with LARP1 binding sites are indicated by red points. (C) MEM-derived sequence motifs identified in LARP1 binding sites on transcript CDS and 3’UTRs under growing and mTOR inactive conditions. (D) Average proportion of guanines present in 3’UTR binding sites compared to the rest of the 3’UTR sequence. (E) Number of human genes under growing or mTOR inactive conditions whose mouse homologs decrease more than 1.5-fold, remain unchanged, or increase more than 1.5- fold in translation efficiency (TE) upon Torin1 treatment. (F) Increased pyrimidine richness at the 3’ ends of RP mRNA 5’UTRs is functionally linked to mTOR-dependent changes in TE rates. (G) LARP1 interacts with ribosomal mRNAs. Ribosomal mRNAs immunoprecipitated by LARP1 IP were quantitated by qPCR.
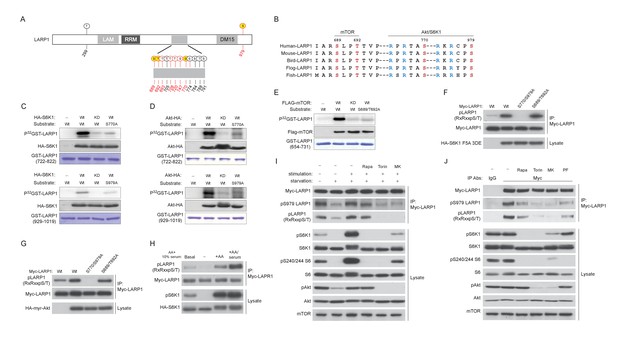
LARP1 is a direct substrate of mTOR, Akt, and S6K1.
(A) Schematic position of LARP1 phosphorylation sites identified by liquid chromatography coupled to electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS). (B) Location and sequence conservation of LARP1 phosphorylation sites. (C–E) S6K1 (C), Akt (D), and mTOR (E) directly phosphorylate LARP1 in vitro. In vitro kinase assay (IVK) were performed with the indicated wild type kinase (WT) and inactive kinase (KD) purified from HEK293T cells using the indicated GST-LARP1 fragments. (F–G) Active S6K1 (F) or Akt (G) enhances phosphorylation of wild-type LARP1 but not the S770A/S979A LARP1 mutant in HEK293T cells. Phosphorylation of LARP1 was detected by phospho-specific-Akt substrate antibody. (H) Levels of LARP1 phosphorylation sites of AGC kinases are enhanced by amino acids or amino acids/growth factors. (I) Amino acids/growth factors-inducible S770/S979 phosphorylation of LARP1 is partially inhibited by rapamycin but largely inhibited by Torin1 or MK-2206. HEK293T cells were serum starved over night and incubated with HBSS with or without the indicated inhibitors for 1 hr before stimulation with DMEM containing 10% FBS for 10 min. (J) Levels of S770/S979 phosphorylation of LARP1 are decreased by rapamycin or S6K1 inhibitor (PF 470861) and further decreased by Torin1 or MK-2206 under steady state growth conditions.
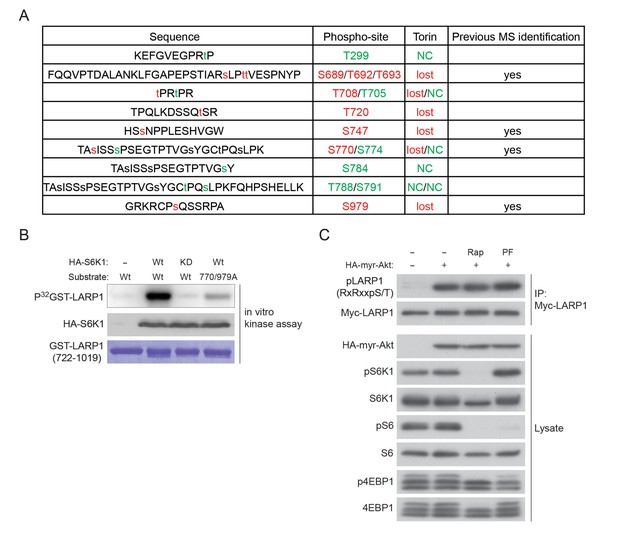
LARP1 is a direct substrate of mTOR, Akt, and S6K1.
(A) A list of LARP1 phosphorylation sites. We performed peptide mapping by liquid chromatography coupled to electrospray ionization tandem mass spectrometry (LC- ESI/MSMS) Qualitative comparisons of resulting phospho-peptides maps revealed that Torin1 treatment abolished phosphorylations of at least eight serine/threonine residues (highlighted by red colure) of LARP1. NC stands for no change. (B) S6K1-dependent phosphorylation of the LARP1 polypeptide (aa 722–1091) containing T770 and S979 is largely decreased by alanine mutations in these residues in vitro. (C) Active Akt is able to phosphorylate LARP1 in HEK293T cells in treated with rapamycin or S6K1 inhibitor (PF 470861).
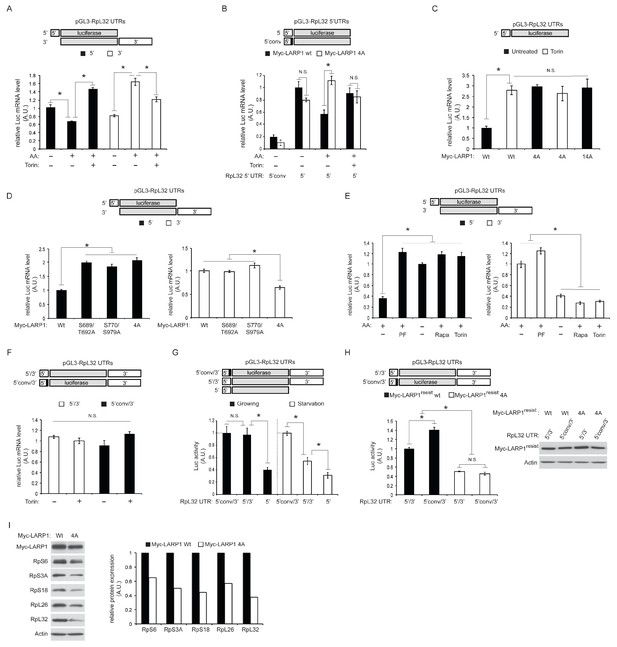
Dynamic rearrangement of LARP1 biding to the UTRs of RpL32 mRNA is regulated by the phosphorylation of LARP1.
(A) The effect of amino acid and mTOR inhibitor on the levels of endogenous LARP1 binding to the 5’ and 3’UTR of RpL32 mRNA. HEK293T cells were transfected with the indicated reporter mRNAs. Endogenous LARP1 was IPed, and the levels of co-IPed luciferase mRNA were determined by qPCR. Data were normalized by input luciferase mRNAs and the amount of IPed LARP1. *p<0.05, mean±SEM (n = 3). (B) LARP1 phosphorylation by mTOR and S6K1/Akt induces its dissociation from the PES in the 5’UTR of RpL32 mRNA. The wild type and the LARP1 4A mutant were transfected with the indicated reporter mRNAs. Data were expressed as Figure 3A. N.S. denotes ‘not significant’. *p<0.05, mean±SEM (n = 3). (C) The effect of alanine substitutions of all the phosphorylation sites of LARP1 identified in this study on the binding to the 5’UTR of RpL32 mRNA under growth conditions. N.S. denotes ‘not significant’. *p<0.05, mean±SEM (n = 3). (D–E) Both mTOR- and S6K1/Akt-dependent LARP1 phosphorylation are necessary for its dissociation from the 5’UTR of RpL32 mRNA, while either mTOR or S6K1/Akt phosphorylation of LARP1 is sufficient to maintain its binding to the 3’UTR. The wild type and the indicated LARP1 mutants were IPed, and the levels of co-IPed 5’ or 3’ reporter mRNA were determined by qPCR (D). HEK293T cells were starved with amino acids or treated with the indicated inhibitors for 1 hr, and levels of co-IPed 5’ or 3’ reporter mRNA with endogenous LARP1 were determined (E). Data were normalized by input luciferase mRNAs and the amount of IPed LARP1. *p<0.05, mean±SEM (n = 3). (F) LARP1 constitutively interacts with the RpL32 reporter RNA containing both the 5’ and 3’ UTRs in a manner independent of the PES in the 5’UTR and mTOR activity. Endogenous LARP1 PAR-CLIP was performed in the presence or absence of Torin1 treatment. Levels of LARP1-bound reporter mRNA were determined by qPCR. Data were normalized by input luciferase mRNAs and the amount of IPed LARP1. N.S. denotes ‘not significant’, mean±SEM (n = 3). (G) The PES motif on 5’UTR of RpL32 is the cis-acting element necessary for translation inhibition in response to mTORC1 inhibition. HEK293T cells were transfected with the indicated reporter mRNAs and grown in normal growth media (10% serum) or mild starvation media (1% serum). After 24 hr, luciferase activity was measured and normalized by luciferase mRNA levels. *p<0.05, mean±SEM (n = 3). (H) Phosphorylation of LARP1 and its dissociation from the 5’UTR are critical for the translation of RpL32. HEK293T cells lacking endogenous LARP1 were transfected with the indicated shRNA-resistant LARPs and reporter mRNAs. 48 hr post-transfection, luciferase activity was measured and normalized by luciferase mRNA levels (left panel). *p<0.05, mean±SEM (n = 3). Levels of transfected Myc-LARP1s were shown (right panel). (I) Phosphorylated LARP1 plays a positive role in ribosomal protein translation. Myc-tagged wild type LARP1 and the 4A mutant LARP1 were stably expressed in HEK293T cells by retrovirus-mediated infection to achieve lower levels of LARP1 expression, and endogenous LARP1 was knockdown by LARP1 shRNA targeting its 5’UTR. Levels of RP proteins were determined by western blotting and the intensity of the bands was quantified.
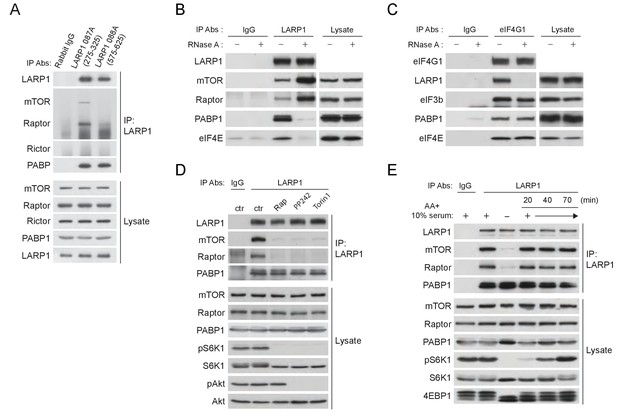
LARP1 recruits mTORC1 to LARP1-interacting mRNPs in a manner dependent of mTORC1 activity.
(A) Endogenous LARP1 co-IPs endogenous mTORC1 in HEK293T cells. LARP1 antibody 087A, but not 088A, co-IPs mTORC1. The LARP1 antibody, 087A or 088A recognizes amino acids 275–325 or 575–625 of LARP1, respectively. (B) The effect of RNAse A on the interaction of LARP1 with mTORC1 and other mRNA binding proteins (PABP1 and eIF4E). Endogenous LARP1 was IPed from the lysates treated with RNAse A. (C) The effect of RNase A on the interaction of LARP1 with the components of the initiation complex. Endogenous eIF4G1 was IPed from the lysates treated with RNAse A. (D) LARP1 interacts with mTORC1 in a manner dependent on mTORC1 activity. (E) LARP1 co-IPs mTORC1 in a growth factor/amino acid stimulation-dependent manner.
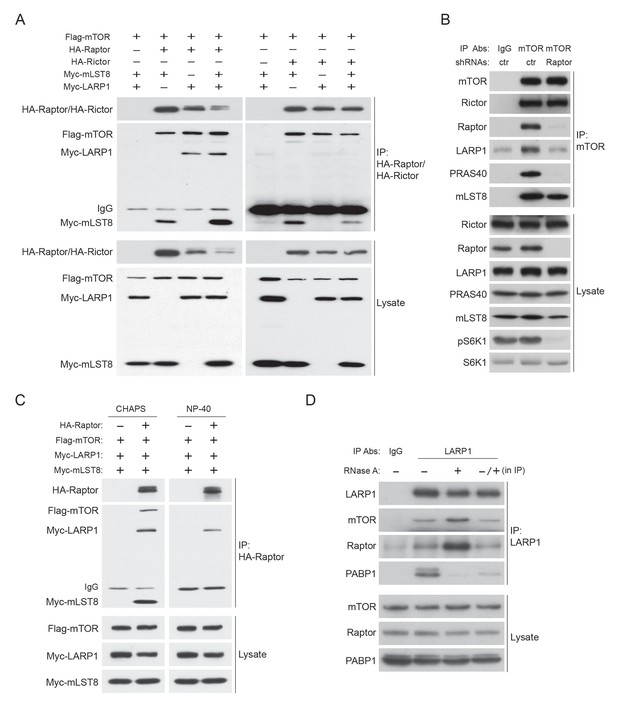
LARP1 interacts with mTORC1 in an RNAse A insensitive manner.
(A) LARP1 interacts with mTORC1. HA-Raptor but not HA-Rictor co-IPs Myc-LARP1 in HEK293T cells. (B) LARP1 interacts with mTORC1 through Raptor. Endogenous mTOR co-IPs endogenous LARP1 in the presence of Raptor. (C) HA-Raptor co-IPs Myc-LARP1 in the presence non-ionic detergent, NP-40, which disrupts the interaction between Raptor and mTOR. (D) PABP1 co-IPed with LARP1 is significantly reduced by RNase A treatment in vitro. The lysates were pre-treated with or without RNase A for one hour and subjected to IP with LARP1 antibody (lane 2 and 3). For lane 4, the LARP1 immunoprecipitant from RNase A untreated lysates was treated with RNase A for one hour. RNase A-treated immunoprecipitant was washed three times with lysis buffer and subjected to western blotting.
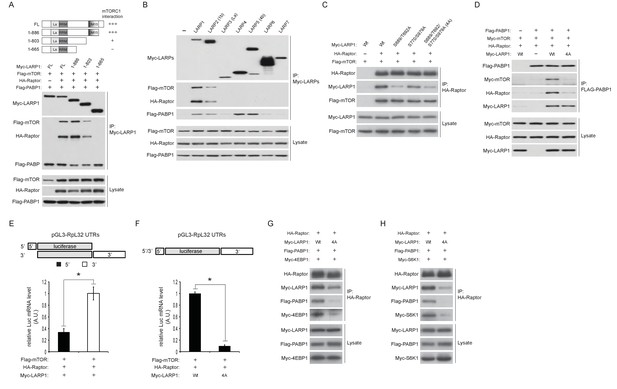
LARP1 scaffolds mTORC1 to LARP1-interacting mRNAs in a manner dependent on LARP1 phosphorylation.
(A) The DM15 domain and its adjacent N-terminal region of LARP1 are required for the interaction with mTORC1. (B) LARP1 and LARP2 but not other LARP family members interact with mTORC1. (C) mTORC1-dependent LARP1 phosphorylation (S689/T692) plays a major role in the interaction between phosphorylated LARP1 and mTORC1. (D) mRNPs containing wild type LARP1 (Wt), but not the phospho-defective LARP1 (4A), associate with mTORC1. (E) mTORC1 preferentially interacts with the 3’UTR than the 5’UTR of RpL32 mRNA. HA-Raptor RIP assays were performed in the presence of wild type LARP1 with the indicated reporter mRNAs. Data were expressed as Figure 3A. (F) mTORC1 interacts with the RpL32 reporter mRNA in the presence of wild type but not LARP1 4A mutant. HA-Raptor RIP assays were performed. (G–H) mTORC1 more interacts with its substrates, 4EBP1 (G) and S6K1 (H) in the presence of wild type LARP1 compared to the LARP1 4A mutant.
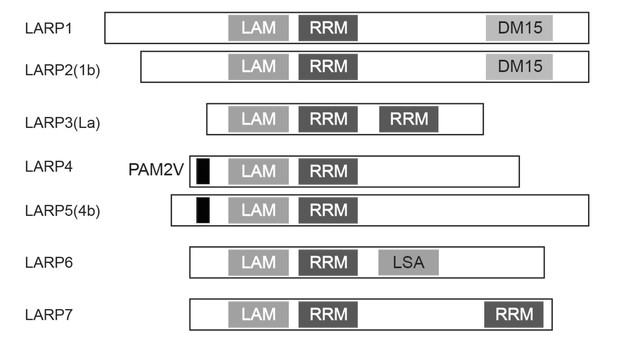
Schematic structure of human LARP family.
All LARP family members possess the La (LAM) and RRM-like (RRM) domains. Only LARP1 and LARP2 have the DM15 repeat domain at their C-terminal regions.
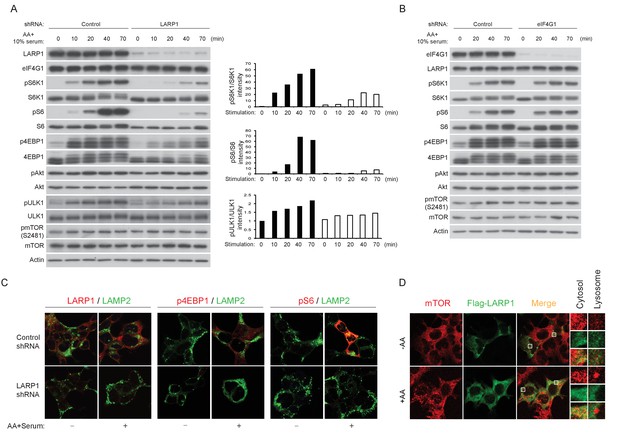
LARP1 enhances mTORC1-dependent phosphorylation of its substrates.
(A–B) LARP1, but not eIF4G1, is required for growth factor/amino acid-induced phosphorylation of mTORC1 substrates in HEK293T cells. (C) LARP1 expresses in cytosolic region and supports growth factor/amino acid-induced S6 and 4EBP1 phosphorylation by mTORC1 in the cytosol. Note that neither LARP1 nor phosphorylated 4EBP1 co-localize with LAMP2, a lysosomal marker. (D) LARP1 co-localizes with mTOR at the cytosolic region in response to amino acid. Note that amino acid-inducible mTOR-positive punctate structures were shown as lysosomes.
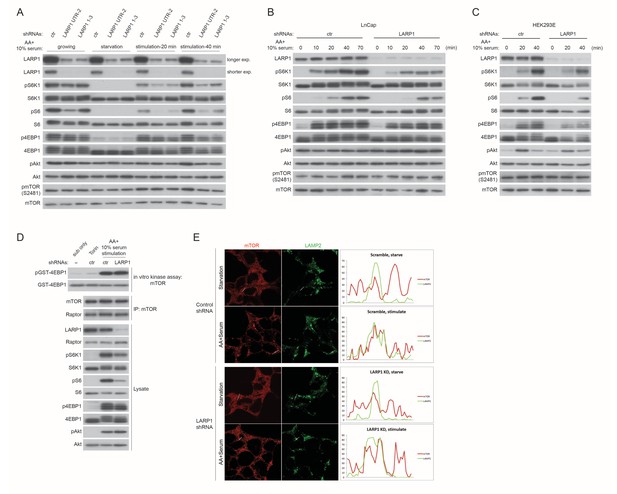
LARP1 enhances mTORC1-dependent phosphorylation of its substrates.
(A) LARP1 knockdown reduces S6K1, S6, and 4EBP1, but not Akt, phosphorylation. LARP1 was knocked-downed with two distinct shRNA constructs (UTR-2 and 1–3), and levels of S6K1, S6, 4EBP1, Akt, and mTOR phosphorylation were monitored under the indicated conditions. (B–C) The effects of LARP1 knockdown on growth factor/amino acid-induced S6K1, S6, 4EBP1, Akt, and mTOR phosphorylation in LnCap and HEK293E (insulin sensitive) cells. (D) The effect of LARP1 knockdown on mTORC1 kinase activity. LARP1 was knock-downed in HEK293T cells, and mTORC1 kinase activity was measured in vitro using recombinant GST-4EBP1 as a substrate. (E) The effect of LARP1 knockdown on growth factor/amino acid-induced mTORC1 localization on lysosomes. HEK203T cells were starved with HBSS for 1 hr and stimulated with DMEM containing 10% FBS. mTOR localizes on lysosomes in response to growth factor/amino acid stimulation under LARP1 knockdown conditions.
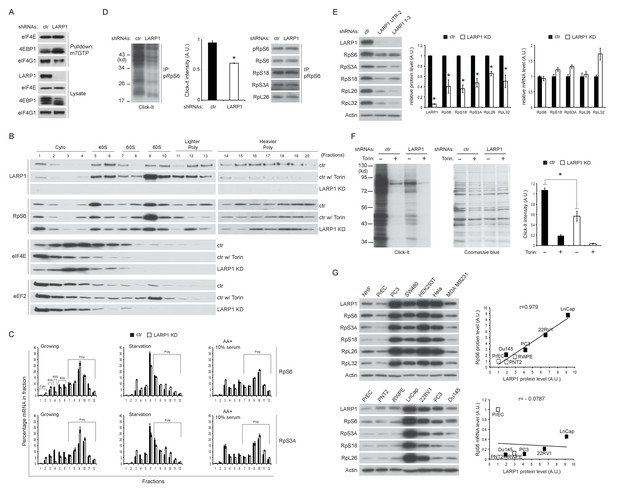
Loss of LARP1 causes defects in the multiple steps of RP mRNA translation.
(A) LARP1 knockdown enhances 4EBP1 binding to the eIF4E precipitated with m7GTP sepharose beads. (B) Loss of LARP1 decreases the expression of eEF2 in the fractions containing active monosomes (80S) and polysomes. (C) RP mRNAs are accumulated in the lighter polysome fractions in LARP1 knockdown cells. (D) Loss of LARP1 decreases the translation of RP mRNAs. Equal amount of ribosomes were immunoprecipitated by phopho-S6 antibody from normal growing HEK293T cells in the presence or absence of LARP1 expression (right panel). Newly synthesized ribosome subunits were visualized (left panel) and quantified (middle panel). *p<0.05, mean±SEM (n = 3). (E) Prolonged LARP1 knockdown (96 hr) decreases the expression of RP proteins (left panels). Levels of RP proteins (middle panel) were quantified and mRNA levels of RP proteins were monitored by qPCR (right panel). Newly synthesized RP proteins were monitored by the Click-It assy *p<0.05 vs control shRNA treatment, mean±SD (n = 3). (F) Prolonged LARP1 knockdown (96 hr) decreases global protein synthesis. p<0.05, mean±SD (n = 3). (left panel). Equal amount of protein loading was visualized by Coomassie blue staining (middle panel). Click-It reaction was quantitated (right panel). *p<0.05 vs control shRNA treatment, mean±SD (n = 3). (G) The expression of LARP1 and RP proteins is enhanced in multiple cancer and transformed cell lines (left panels). Correlation between LARP1 protein vs. RpS6 protein (right upper) or RpS6 mRNA (right lower) in prostate epithelial cells. Open or close square indicates normal or benign prostate epithelial cells or metastatic prostate cancer cells, respectively.
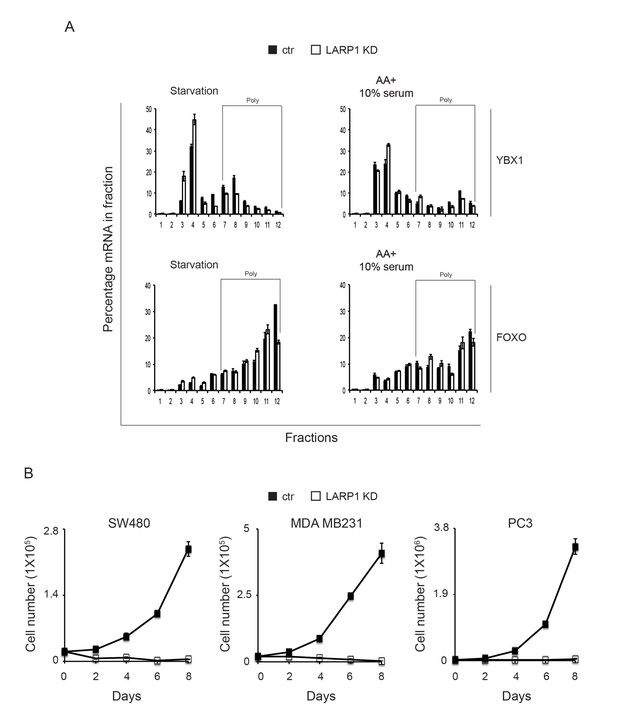
Loss of LARP1 causes defects in the multiple steps of RP mRNA translation.
(A) The effect of LARP1 knockdown on the distribution of LARP1-non-interacting mRNAs in the polysome fractions. HEK293T cells treated with control or LARP1 shRNA were subjected to polysome fractionation. Levels of the indicated mRNAs in the fractions were determined by qPCR. Data were expressed as a percentage of the indicated mRNAs in each fraction. (B) The effect of LARP1 knockdown on the proliferation of cancer cells. SW480, MDA MB231, and PC3 cells were treated with control or LARP1 shRNA. 48 hr post shRNA infection, the same number of the cancer cells treated with control or LARP1 shRNA was seeded and changes of cell number were monitored.
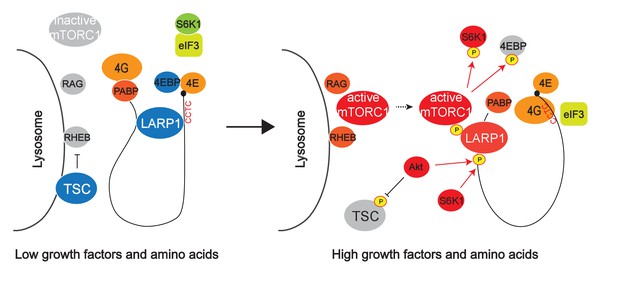
Hypothetical model for mTORC1-dependent LARP1-interacting RP mRNA translation.
mTOR Complex 1 (mTORC1) is activated by growth factors and amino acids on the lysosomal surface. After its activation, mTORC1 is recruited to cytoplasmic mRNPs via unknown mechanisms. LARP1, which stably interacts with RP mRNAs, is phosphorylated by mTORC1, Akt, and S6K1. These phosphorylations induce the dissociation of LARP1 from the PES in the 5’UTRs of RP mRNAs and relieve its inhibitory effect on the translation. Concomitantly, the phosphorylated LARP1 that strongly interact with the 3’UTR of RP mRNAs scaffolds mTORC1 on translationally-competent RP mRNAs and supports mTORC1-dependent phosphorylation of S6K1 and 4EBP1 thereby enhancing translation initiation and elongation of LARP1-interacting mRNAs.
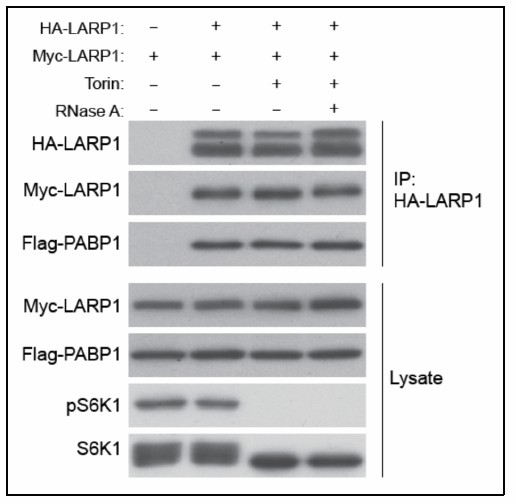
LARP1 forms at least a dimer.
HEK293T cells co-transfected with HA- and MYC-LARP1 were treated with Torin1 for 1 hour before harvest. The lysate was treated with RNAseA prior to IP (lane 4).
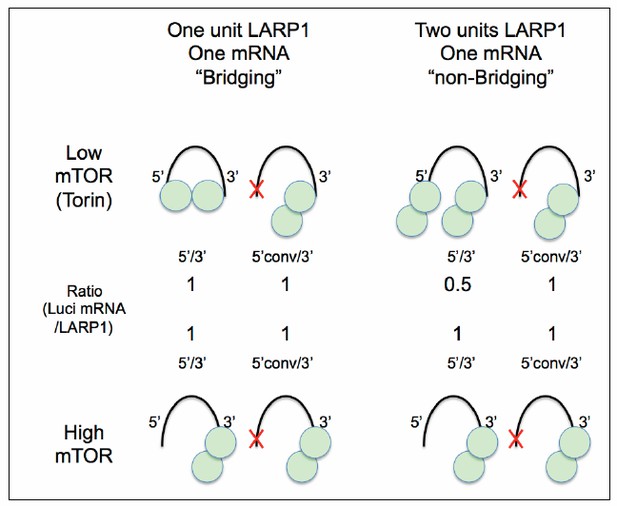
A Hypothetical model for LARP1-RpL32 configuration.
The data presented in this study support “One unit LARP1-one mRNA bridging” configuration under low mTOR activity conditions (left).
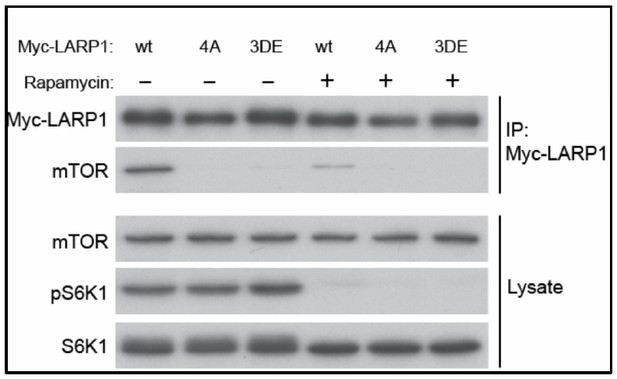
Wild type but not phopho-defectitve or phospho-mimetic LARP1 interacts with mTOR in a rapamycin sensitive manner.
https://doi.org/10.7554/eLife.25237.022Additional files
-
Supplementary file 1
LARP1 binding sites.
- https://doi.org/10.7554/eLife.25237.016
-
Supplementary file 2
Gene ontology terms enriched in LARP1-bound genes under growing and mTOR inactive conditions.
- https://doi.org/10.7554/eLife.25237.017
-
Supplementary file 3
LARP1-bound genes classified as non-TR, TR, and RP-encoding genes.
- https://doi.org/10.7554/eLife.25237.018
-
Supplementary file 4
Location of 5’TOP/5’TOP-like and LARP1 binding site in select 5’UTRs.
- https://doi.org/10.7554/eLife.25237.019





