Parallel Activin and BMP signaling coordinates R7/R8 photoreceptor subtype pairing in the stochastic Drosophila retina
Figures
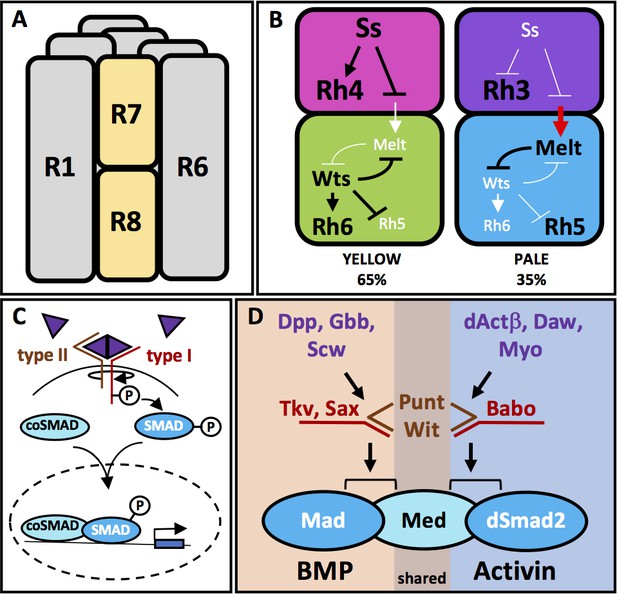
Ommatidia architecture and the mechanisms of TGFβ pathway signaling.
(A) The six outer photoreceptors R1-R6 all express Rh1 and are involved in motion detection and image formation in dim light. They surround the rhabdomeres of the two inner photoreceptors R7 and R8, which are located on top of each other, thus sharing the same optic path. (B) The transcription factor Ss is expressed in 65% of R7 cells (yellow cells), activating Rh4 and repressing Rh3. Ss inhibits the instructive signal from R7 to R8, allowing for the default phosphorylation and activation of Wts in R8 and subsequent expression of Rh6 and repression of Rh5. Wts makes up one half of a double-negative feedback loop that establishes and maintains R8 subtypes. The other half of the loop, Melt, is expressed in R8s downstream of the other 35% of R7 cells (pale cells). These pale R7 cells lack Ss expression, allowing for expression of Rh3 and instructive signaling to R8 (red arrow). The instructive signal likely activates Melt, which represses Wts, allowing for Rh5 expression. (C) TGFβ superfamily ligands induce oligomerization of Type I and Type II serine-threonine kinase receptors. Binding of the ligand dimer to the Type II receptor initiates its kinase activity, phosphorylating residues on the Type I receptor, which becomes activated. Type I receptors then phosphorylate members of the receptor regulated (R)-SMAD family of transcription factors, allowing them to bind co-SMADs, translocate to the nucleus and activate or repress transcription of downstream target genes. (D) The TGFβ pathway in Drosophila contains both BMP and Activin subfamilies. The BMP subfamily is composed of three ligands, Dpp, Gbb and Scw, two Type I receptors, Tkv and Sax and one R-SMAD, Mad. The Activin subfamily also contains three ligands, dActβ, Daw and Myo, but only one Type I receptor, Babo and one R-SMAD, dSmad2. Both subfamilies share the Type II receptors Punt and Wit as well as the Co-SMAD, Med.
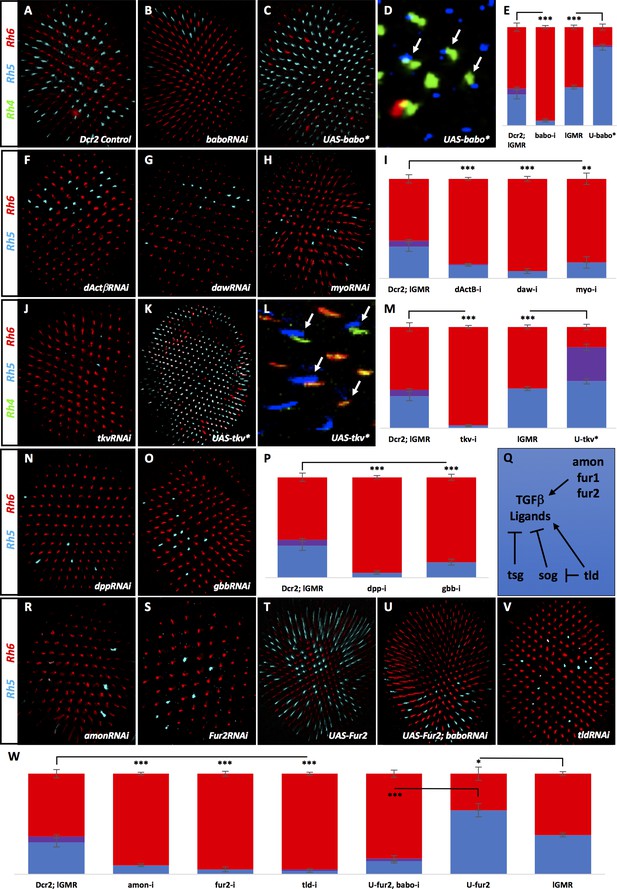
Many TGFβ ligands and receptors are required non-redundantly to specify R8 subtypes.
Graph Legend: Red is the percentage of R8 photoreceptors that expressed Rh6, Blue is the percentage that expressed Rh5 and Purple is Co-expression of Rh5 + Rh6. Error bars report standard error of the mean. Paired t-tests were performed against percentages of Rh5 expression and significance is denoted where applicable (***=p≤0.01, **=p≤0.05, *=p≤0.1, ns = not significant). Where significance (or non-significance) is noted, follow the horizontal bar above the significance value to its termination in a vertical tick to find the comparative genotype. Percentage values for each genotype are listed as (Rh5/(Rh5 + Rh6)/Rh6). (A) lGMR-Gal4 > UAS-Dcr2 control retinas (32/6/62) (n = 6). Antibodies label Rh5 in blue and Rh6 in red. (B) lGMR-Gal4 > UAS-Dcr2; UAS-BaboRNAi drastically reduced pale subtypes (5/0/95) (n = 13). (C) lGMR-Gal4 > UAS-Babo* increased pale subtypes (80/2/19) (n = 20). (D) lGMR-Gal4 > UAS-Babo* retinas altered subtype ratios of R8 cells without altering R7, resulting in mispairing of yellow, Rh4-expressing R7 cells (green) and pale, Rh5-expressing R8 cells (blue) (arrows). (E) Quantification of experiments A-C with the addition of a lGMR-Gal4 control (39/0/61) (n = 7). (F) Using lGMR-Gal4 to express UAS-Dcr2 and UAS-dActβRNAi (14/0/86) (n = 5), UAS-DawRNAi (7/0/93) (n = 10) (G), and UAS-MyoRNAi (16/0/84) (n = 5) (H) decreased pale subtypes. (I) Quantification for F-H. (J) lGMR > UAS-Dcr2; UAS-TkvRNAi (3/0/97) (n = 6) also reduced pale R8 subtypes. (K) lGMR-Gal4 >UAS-Tkv* increased pale subtypes (46/34/20) (n = 16). (L) lGMR-Gal4 >UAS-Tkv* retinas altered the subtype ratios of R8 cells without altering R7, resulting in mispairing of yellow, Rh4-expressing R7 cells and pale, Rh5-expressing R8 cells (arrows). Some Rh5 +Rh6 co-expression occurred in UAS-Tkv* retinas (red + blue staining). (M) Quantification of J-K. (N) lGMR-Gal4 > UAS-Dcr2 and UAS-DppRNAi (5/0/95) (n = 5) or UAS-GbbRNAi (15/0/85) (n = 9) (O) reduced pale subtypes. (P) Quantification of N-O. (Q) Schematic of TGFβ ligand regulation: Amon, Fur1 and Fur2 are furin-type proprotein convertases capable of cleaving and activating ligands. Tsg and Sog bind ligands and prevents their activity while Tld can cleave Sog to undue repression or cleave ligands directly to activate them. (R) lGMR-Gal4 > UAS-Dcr2 with AmonRNAi (9/0/91) (n = 4) or Fur2RNAi (5/0/95) (n = 4) (S) reduced pale subtypes. (T) lGMR-Gal4 > UAS-Fur2 increased pale subtypes (64/0/36) (n = 3). (U) Removing babo prevented UAS-Fur2 from increasing pale subtypes (lGMR-Gal4 > UAS-Dcr2; UAS-BaboRNAi; UAS-Fur2) (13/3/84) (n = 6). (V) lGMR-Gal4 > UAS-Dcr2; UAS-TldRNAi (5/0/95) (n = 3) reduced pale subtypes. (W) Quantification of R-V.
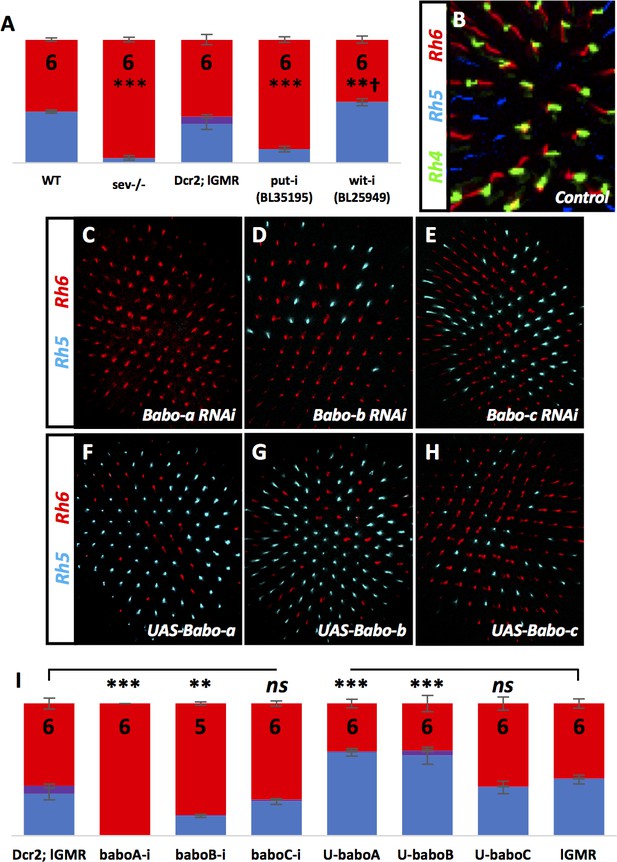
Babo-a, -b, and Put but not Babo-c or Wit are required for R8 subtypes.
Graph Legend: Red is the percentage of R8 photoreceptors that expressed Rh6, Blue is the percentage that expressed Rh5 and Purple is Co-expression of Rh5 + Rh6. Error bars report standard error of the mean. Paired t-tests were performed against percentages of Rh5 expression and significance is denoted where applicable (***=p≤0.01, **=p≤0.05). † denotes a change in Rh5/Rh6 ratio that was significant but in the unexpected direction. N values listed on bar graph. Percentage values for each genotype are listed as (Rh5/(Rh5 + Rh6)/Rh6). (A) sev14 mutants in a WT background drastically decreased the number of pale cells to 4% (4/0/96) - compare to WT control (42/0/58). Removing put resulted in decreased pale subtypes (22/2/75) similar to removing the Type I receptors tkv or babo while the Type II receptor, Wit, was not required for the pale subtype (49/0/51) – compare to Dcr2; lGMR control (32/6/62). (B) WT retinas pair Rh4-expressing R7 cells (green) with Rh6-expressing R8 cells (red). (C) lGMR-Gal4 > UAS-Dcr2; UAS-Babo-aRNAi completely eliminated pale subtypes (0/0/100). (D) lGMR-Gal4 > UAS-Dcr2; UAS-Babo-bRNAi also reduced pale subtypes (15/1/84), however lGMR-Gal4 > UAS-Dcr2; UAS-Babo-cRNAi did not reduce pale subtypes (26/1/73) (E). (F) Overexpressing Babo-a (lGMR-Gal4 >UAS Babo-a) (63/1/36) and Babo-b (lGMR-Gal4 > UAS Babo-b) (61/4/36) (G) increased pale subtypes. (H) Overexpressing Babo-c (lGMR-Gal4 > Babo c) did not alter the ratio of pale/yellow subtypes (37/0/63). (I) Quantification of C-H.
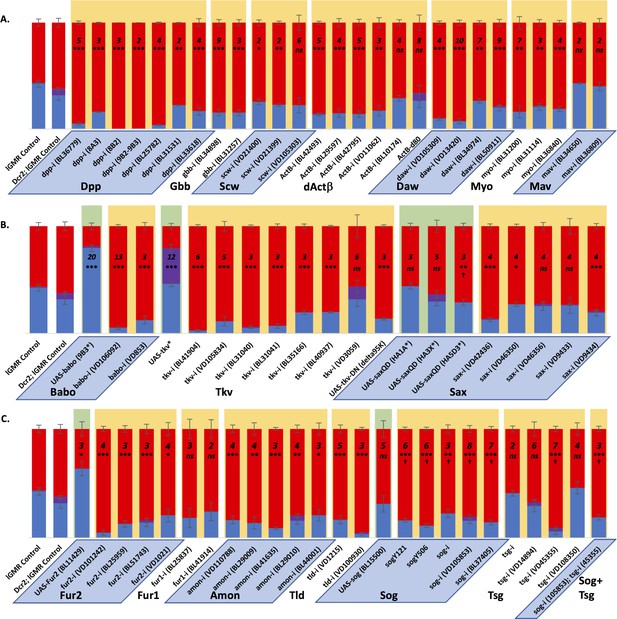
Result validation using additional alleles.
Graph Legend: Red represents the percentage of R8 photoreceptors that expressed Rh6, Blue is the percentage that expressed Rh5 and purple is co-expression of Rh5 + Rh6. Error bars report standard error of the ,ean. Paired t-tests were performed against percentages of Rh5 expression and significance is denoted where applicable (***=p≤0.01, **=p≤0.05, *=p≤0.1, ns = not significant). For each graph, color coding has been added for ease of reading. Results backed with green are overexpression constructs, those backed with gold are downregulating constructs. Alternating, bottom-labeled parallelograms of blue or white separate groups of genes tested. Significance is listed for each construct relative to the appropriate control at the extreme left of each graph; lGMR-Gal4 Control for green-backed genotypes, lGMR-Gal4 >UAS-Dcr2 Control for gold-backed genotypes. N values listed above significance. † denotes a change in Rh5/Rh6 ratio that was significant but in the unexpected direction. Due to some level of variability in Rh5/Rh6 ratios within distinct genetic backgrounds, we imposed a cutoff for including factors as part of the pale subtype instructive signal; the requirement we set was that greater than 50% of misexpression constructs presented a phenotype and that at least one of those was highly significant (p≤0.01). (A) We used a number of RNAi and mutant constructs to confirm our TGFβ ligand results. (B) We also used a number of RNAi, Dominant negative and overexpression constructs to confirm our TGFβ receptor results. (C) We confirmed our processing factor results with additional RNAi and overexpression constructs. Of note, removing sog, in all cases, significantly increased yellow subtypes, opposite to our predictions. It is possible that Dpp requires Sog – as it does in the embryo to diffuse over long distances (Decotto and Ferguson, 2001) – to reach receptors on the R8 cell, even though the distance to travel is short. Therefore, removing sog would prevent Dpp activity as would removing tld, which would be required to cleave the Sog/Dpp complex for Dpp activation of Tkv. Tsg appears dispensable in this role in the retina.
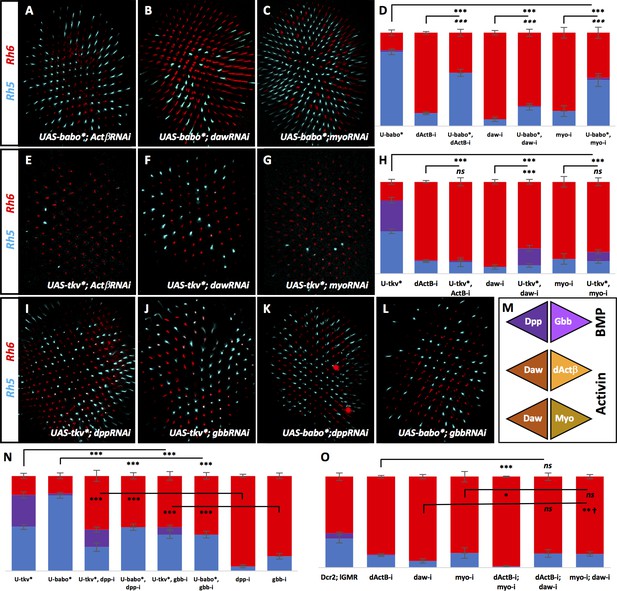
BMP and Activin ligands signal to their canonical receptors.
Graph Legend: Red is the percentage of R8 photoreceptors that expressed Rh6, Blue is the percentage that expressed Rh5 and Purple is Co-expression of Rh5 + Rh6. Error bars report standard error of the mean. Paired t-tests were performed against percentages of Rh5 expression and significance is denoted where applicable (***=p≤0.01, **=p≤0.05, *=p≤0.1, ns = not significant). Where significance (or non-significance) is noted, follow the horizontal bar above the significance value to its termination in a vertical tick to find the comparative genotype. Percentage values for each genotype are listed as (Rh5/(Rh5 + Rh6)/Rh6). (A) Expressing UAS-Babo* rescued some yellow subtypes induced by removing dActβ (lGMR-Gal4 > UAS-Dcr2; UAS-Babo*; UAS-dActβRNAi) (57/0/43) (n = 6), daw (to a lesser extent) (lGMR-Gal4 > UAS-Dcr2; UAS-Babo*; UAS-DawRNAi) (21/1/78) (n = 23) (B) and myo (lGMR-Gal4 >UAS-Dcr2; UAS-Babo*; UAS-MyoRNAi) (49/2/48) (n = 6) (C). (D) Quantification of A-C. (E) Expressing UAS-Tkv* could not rescue yellow subtypes induced by removing dActβ (lGMR-Gal4 > UAS-Dcr2; UAS-Tkv*; UAS-dActβRNAi) (12/1/86) (n = 6). (F) Tkv* did slightly, yet significantly, rescue loss of daw (lGMR-Gal4 > UAS-Dcr2; UAS-Tkv*; UAS-DawRNAi) (9/18/72) (n = 13) but could not rescue loss of myo (lGMR-Gal4 > UAS-Dcr2; UAS-Tkv*; UAS-MyoRNAi) (14/10/76) (n = 8) (G). (H) Quantification of E-G. (I) Expressing UAS-Tkv* rescued many yellow subtypes induced by removing dpp (lGMR-Gal4 > UAS-Dcr2; UAS-Tkv*; UAS-DppRNAi) (25/18/56) (n = 15) or gbb (lGMR-Gal4 > UAS-Dcr2; UAS-Tkv*; UAS-GbbRNAi) (38/8/54) (n = 12) (J). (K) Expressing UAS-Babo* also rescued many yellow subtypes induced by removing dpp (lGMR-Gal4 > UAS-Dcr2; UAS-Babo*; UAS-DppRNAi) (46/0/54) (n = 7) or gbb (lGMR-Gal4 > UAS-Dcr2; UAS-Babo*; UAS-GbbRNAi) (38/0/62) (n = 6) (L). (M) Schematic for hypothetical ligand dimerization possibilities upstream of Tkv and Babo activation. Dpp + Gbb may heterodimerize to activate Tkv while Daw + dActβ and Daw + Myo may heterodimerize and signal at highly regulated levels for Babo activation. (N) Quantification of I-L. (O) Co-expression of dActβRNAi and MyoRNAi (lGMR-Gal4 > UAS-Dcr2; UAS-dActβRNAi; UAS-MyoRNAi) (1/0/99) (n = 6) exacerbated specification of yellow subtypes induced in dActβRNAi or MyoRNAi retinas alone. However, combining DawRNAi with either dActβRNAi (lGMR-Gal4 > UAS-Dcr2; UAS-dActβRNAi; UAS-DawRNAi) (15/0/85) (n = 7) or MyoRNAi (lGMR-Gal4 > UAS-Dcr2; UAS-MyoRNAi; UAS-DawRNAi) (15/0/85) (n = 12) could not exaggerate the phenotypes of any of the three alone. † denotes a change in Rh5/Rh6 ratio that was significant but in the unexpected direction.
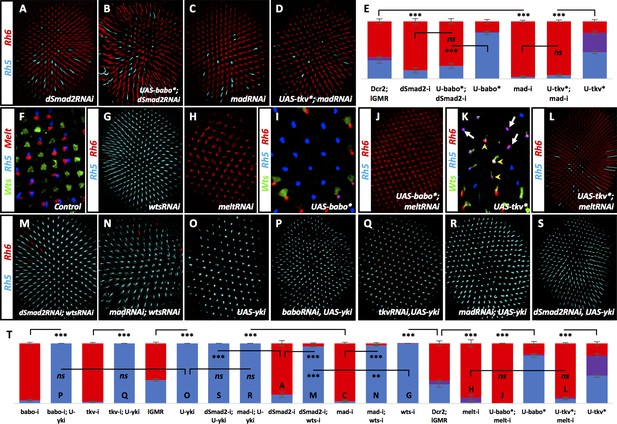
Babo, through dSmad2 and Tkv, through Mad, regulate Wts and Melt.
Graph Legend: Red is the percentage of R8 photoreceptors that expressed Rh6, Blue is the percentage that expressed Rh5 and Purple is Co-expression of Rh5 +Rh6. Error bars report standard error of the mean. Paired t-tests were performed against percentages of Rh5 expression and significance is denoted where applicable (***=p ≤ 0.01, **=p ≤ 0.05, ns = not significant). Where significance (or non-significance) is noted, follow the horizontal bar above the significance value to its termination in a vertical tick to find the comparative genotype. Percentage values for each genotype are listed as (Rh5/(Rh5 +Rh6)/Rh6). (A) Expressing UAS-Dcr2 and UAS-dSmad2RNAi with lGMR-Gal4 reduced pale subtypes (lGMR-Gal4 >UAS-Dcr2; UAS-dSmad2RNAi) (15/0/85) (n = 4) and this reduction was maintained even when UAS-Babo* was co-expressed (lGMR-Gal4 >UAS-Dcr2; UAS-dSmad2RNAi; UAS-Babo*) (23/0/77) (n = 7) (B). (C) Expressing UAS-Dcr2 and UAS-madRNAi with lGMR-Gal4 also reduced pale subtypes (lGMR-Gal4 >UAS-Dcr2; UAS-MadRNAi) (4/0/96) (n = 7). (D) Expressing UAS-Tkv* could not rescue MadRNAi-induced loss of pale subtypes (lGMR-Gal4 >UAS-Dcr2; UAS-MadRNAi, UAS-Tkv*) (6/0/94) (n = 9). (E) Quantification of A-D. (F) Wild-type retinas express Melt (red) and Wts (green) in mutually exclusive subsets of R8 cells; Melt (red) with Rh5 (blue) in the pale subset and Wts (green) with Rh6 (not shown) in the alternate, yellow R8 subset. (G) Removing wts (lGMR-Gal4 > UAS-Dcr2; UAS-WtsRNAi) (100/0/0) (n = 8) led to total loss of yellow subtype while removing melt (lGMR-Gal4 > UAS-Dcr2; UAS-MeltRNAi) (2/8/90) (n = 5) resulted in the opposite, loss of pale subtypes (H). (I) Expressing Babo* increased pale subtypes (blue), which was concomitant with a loss of Wts expression (green). (J) Removing melt in retinas that overexpress Babo* (lGMR-Gal4 > UAS-Dcr2; UAS-Babo*; UAS-MeltRNAi) (0/0/100) (n = 11) prevented Babo*-induced pale subtypes. (K) Overexpression of Tkv* increased the number of ommatidia expressing Rh5 (blue) in R8. In this genotype, increased Rh5 expression was often co-expressed with Rh6 (magenta). Increased Rh5 expression was coincident with loss of Wts, even in some instances where Rh6 was still present (white arrows); however, Wts remained in other R8 cells that co-expressed Rh5 + Rh6 (yellow arrowheads). (L) Removing melt from Tkv* retinas (lGMR-Gal4 > UAS-Dcr2; UAS-Tkv*; UAS-MeltRNAi) (7/3/90) (n = 46) resulted in a loss of Tkv*-induced Rh5 expression. (M) While significantly different from wtsRNAi controls (due to extremely low variance), removing wts prevented increased yellow subtypes seen in dSmad2RNAi (lGMR-Gal4 > UAS-Dcr2; UAS-dSmad2RNAi; UAS-WtsRNAi) (95/0/5) (n = 6) and MadRNAi (lGMR-Gal4 > UAS-Dcr2; UAS-MadRNAi; UAS-WtsRNAi) (97/0/3) (n = 12) (N) retinas. (O) lGMR-Gal4 > UAS Yki retinas exhibited 100% pale subtypes (100/0/0) (n = 7). (P) Expressing UAS-Yki overcame the high incident of yellow subtypes induced by expressing BaboRNAi (lGMR-Gal4 > UAS-Dcr2; UAS-BaboRNAi; UAS-Yki) (100/0/0) (n = 6), TkvRNAi (lGMR-Gal4 > UAS-Dcr2; UAS-TkvRNAi; UAS-Yki) (100/0/0) (n = 6) (Q), MadRNAi (lGMR-Gal4 >UAS-Dcr2; UAS-MadRNAi; UAS-Yki) (100/0/0) (n = 6) (R) and dSmad2RNAi (lGMR-Gal4 >UAS-Dcr2; UAS-dSmad2RNAi; UAS-Yki) (100/0/0) (n = 6) (S). (T) Quantification of G-H, J, L-S. Letters (P, Q, O…) have been added to applicable bars to help identify corresponding image panels.
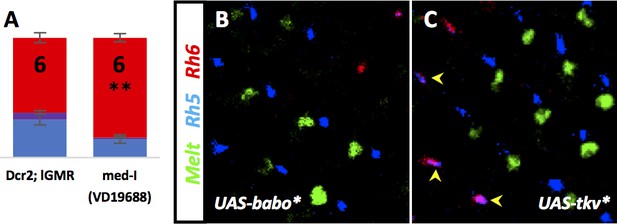
Med, Babo, and Tkv are required for R8 subtypes.
Graph Legend: Red is the percentage of R8 photoreceptors that expressed Rh6, Blue is the percentage that expressed Rh5 and Purple is Co-expression of Rh5 + Rh6. Error bars report standard error of the mean. Paired t-tests were performed against percentages of Rh5 expression and significance is denoted where applicable (**=p≤0.05). N values are listed on the graph. Percentage values for each genotype are listed as (Rh5/(Rh5 + Rh6)/Rh6). (A) Removing the co-SMAD med resulted in decreased pale subtypes (15/1/83) similar to removing its binding partners mad or dSmad2 – compare to Dcr2; lGMR control (32/6/62). (B) Overexpressing Babo* using lGMR-Gal4 lead to expression of Melt (green) and Rh5 (blue). (C) Overexpression of Tkv* using lGMR-Gal4 also lead to expression of Melt (green) and Rh5 (blue), although in some cases where Rh5 and Rh6 become co-expressed (magenta), Melt expression remained absent (yellow arrowheads). These are likely the same ommatidia that retain wts expression (see Figure 4K).
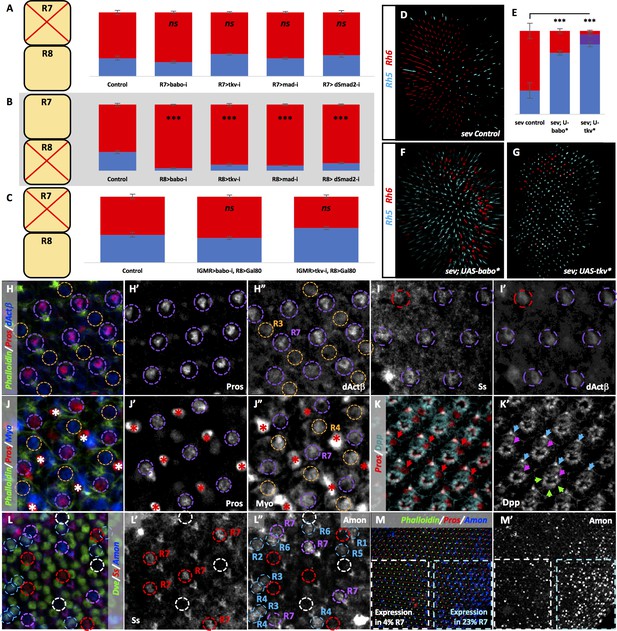
TGFβ ligands are processed specifically in pale R7 and activate their receptors and subsequent R-SMADs in R8.
Graph Legend: Red is the percentage of R8 photoreceptors that expressed Rh6, Blue is the percentage that expressed Rh5 and Purple is Co-expression of Rh5 +Rh6. Error bars report standard error of the mean. Paired t-tests were performed against percentages of Rh5 expression and significance is denoted where applicable (***=p ≤ 0.01, ns = not significant). Percentage values for each genotype are listed as (Rh5/(Rh5 +Rh6)/Rh6). Control for A is PM1.81-Gal4; Pros-Gal4 >UAS-Dcr2 (29/0/71) (n = 6). For B, the control is Sca-Gal4; Sens-Gal4 > UAS-Dcr2 (28/0/72) (n = 7). Control for C is lGMR-Gal4 > UAS-Dcr2; Sca-lexA > lexAOP-Gal80 (42/0/58) (n = 7). (A) Removing babo (PM1.81-Gal4; Pros-Gal4 > UAS-Dcr2; UAS-BaboRNAi) (23/0/77) (n = 6), tkv (PM1.81-Gal4; Pros-Gal4 > UAS-Dcr2; UAS-TkvRNAi) (35/0/65) (n = 7), mad (PM1.81-Gal4; Pros-Gal4 >UAS-Dcr2; UAS-MadRNAi) (28/0/72) (n = 5), or dSmad2 (PM1.81-Gal4; Pros-Gal4 >UAS-Dcr2; UAS-dSmad2RNAi) (33/0/67) (n = 6) specifically in R7 had no effect on Rh5/Rh6 ratios. (B) Removing babo (Sca-Gal4; Sens-Gal4 > UAS-Dcr2; UAS-BaboRNAi) (4/0/96) (n = 6), tkv (Sca-Gal4; Sens-Gal4 > UAS-Dcr2; UAS-TkvRNAi) (10/0/90) (n = 7), mad (Sca-Gal4; Sens-Gal4 > UAS-Dcr2; UAS-MadRNAi) (8/0/92) (n = 4), or dSmad2 (Sca-Gal4; Sens-Gal4 > UAS-Dcr2; UAS-dSmad2RNAi) (12/0/88) (n = 4) specifically in R8 reduced the number of pale subtypes as we saw with lGMR-Gal4 drivers. (C) Silencing Gal4 expression specifically in R8 attenuated the lGMR-Gal4 > UAS BaboRNAi- and TkvRNAi-induction of yellow R8 subtypes (lGMR-Gal4 > UAS-Dcr2; UAS-BaboRNAi; Sca-lexA > lexAOP-Gal80) (37/0/63) (n = 6) (lGMR-Gal4 > UAS-Dcr2; UAS-TkvRNAi; Sca-lexA > lexAOP-Gal80) (52/0/48) (n = 4). (D) sev mutations genetically ablate R7 cells resulting in increased amounts of default yellow R8 cells. Our sev mutant control, which carries a lGMR-Gal4 driver, expressed higher levels of pale subtypes (29/0/71) than observed in sev mutants in a WT background (4/0/96), likely due to genetic background (n = 3). (E) Quantification of D, F-G. (F) Expressing UAS-Babo* in a sev mutant (sev14; lGMR-Gal4 > UAS-Babo*) (74/0/26) (n = 5) increased the number of pale subtypes significantly. (G) Expressing UAS-Tkv* in a sev mutant (sev14; lGMR-Gal4 > UAS-Tkv*) (84/12/4) (n = 5) increased pale subtypes significantly. (H-M) Pupal retinas, 45–55 hr APF. (H) dActβ was expressed in all R7 cells (circled in purple) and less strongly in some R3 cells (circled in orange). (H’) Single-channel staining of the Prospero (Pros) transcription factor, which is expressed in all R7 cells. (H’). Single-channel dActβ protein reporter overlaped with Pros expression. (I) The Spineless (Ss) antibody marks yellow R7 cells (circled in purple). (I’) dActβ was expressed in both Ss-positive yellow R7 cells (circled in purple) and Ss-negative pale R7 cells (circled in red). (J) A Myo protein reporter was expressed in all R7 cells (circled in purple) as well as R4 cells (circled in orange). Bristle cells also labeled brightly (marked with asterisks). (J’) Single-channel Pros staining. (J’) Single-channel Myo reporter. (K) A Dpp-GFP transcriptional reporter was expressed in all photoreceptors. Pros marks R7 cells (red arrows). (K’) Single-channel Dpp reporter. Blue arrows mark R6 expression; green arrows mark expression in photoreceptors 1–5; pink arrows mark expression in R7. (L) An Amon-Gal4 transcriptional reporter was expressed in some pale R7 cells (circled in purple) but is absent in others (circled in white). It was absent from yellow R7 cells (circled in red). The reporter was also expressed in some outer photoreceptors (circled in blue). (L’) Single channel Ss staining marks yellow R7 cells (circled in red). (L’) Single-channel Amon reporter. Amon pale R7 expression – 9%, yellow R7 expression – 1%. N = 2678 ommatidia. (M) Amon expression was variable across individual pupal retina. (M’) Single channel Amon reporter expression.
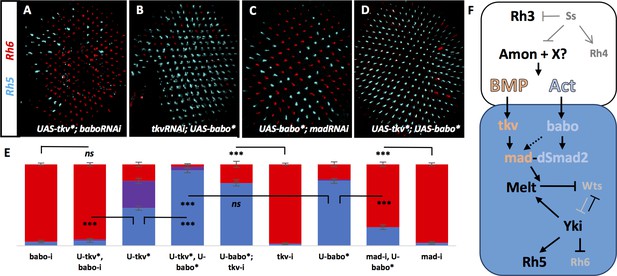
Tkv and Babo signal in parallel to establish a ‘double-checked’ pathway, ensuring correct R7/R8 pairing.
Graph Legend: Red is the percentage of R8 photoreceptors that expressed Rh6, Blue is the percentage that expressed Rh5 and Purple is Co-expression of Rh5 + Rh6. Error bars report standard error of the mean. Paired t-tests were performed against percentages of Rh5 expression and significance is denoted where applicable (***=p ≤ 0.01, ns = not significant). Where significance (or non-significance) is noted, follow the horizontal bar above the significance value to its termination in a vertical tick to find the comparative genotype. Percentage values for each genotype are listed as (Rh5/(Rh5 + Rh6)/Rh6). (A) Removing babo resulted in a high incidence of yellow subtypes even when UAS-Tkv* was expressed (lGMR-Gal4 > UAS-Dcr2; UAS-BaboRNAi; UAS-Tkv*) (6/2/92) (n = 6). (B) Removing tkv could not eliminate increased pale subtypes in UAS-Babo* retinas (lGMR-Gal4 > UAS-Dcr2; UAS-TkvRNAi; UAS-Babo*) (77/0/23) (n = 9). (C) Removing mad significantly reduced pale subtypes induced by expressing UAS-Babo* (lGMR-Gal4 > UAS-Dcr2; UAS-MadRNAi; UAS-Babo*) (23/0/77) (n = 15). (D) Expressing both UAS-Tkv* and UAS-Babo* (lGMR-Gal4 > UAS-Tkv*; UAS-Babo*) (93/5/3) (n = 7) significantly increased pale subtypes above that induced by either alone. (E) Quantification of A-D. (F) Model. Lack of ss in pale R7 cells allows for expression of Amon and possibly other proprotein convertases to activate signaling of BMP and Activin ligands, which activate canonical receptors and R-SMADs Tkv/Mad and Babo/dSmad2, respectively, in R8. Cooperative action of Mad and dSmad2 then regulate the Melt/Wts bi-stable loop, allowing Yki/Sd to induce expression of Rh5 and repression of Rh6.
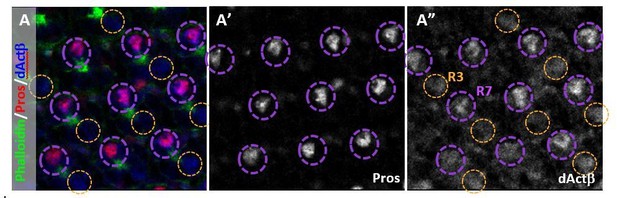
(A) dActβ co-localized with the R7 marker Prospero (Pros) (circled in purple).
It also weakly labeled some R3 cells. (A’) Pros marks R7 cells. (A”) dActβ was expressed in R7 and R3.
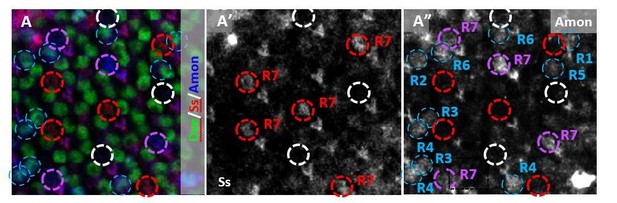
(A) Amon was expressed in pale R7 cells (circled in purple) but not in yellow R7s (circled in red).
It was also expressed in some outer PRs (circled in blue) and absent from other pale R7 cells (circled in white). (A’) Ss marks yellow R7 cells (circled in red). (A”) Amon was present in some but not all Ss-negative pale R7s (circled in purple) but absent from Ss-positive yellow R7s (circled in red).
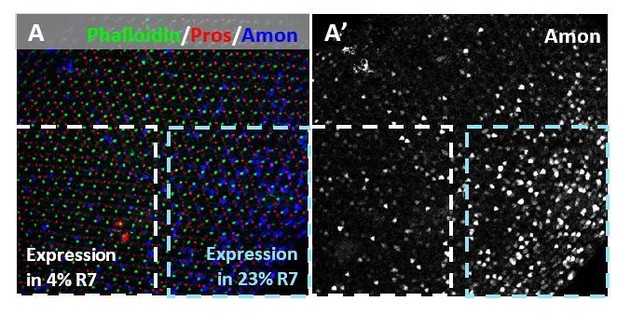
Across a single pupal retina, Amon expression varied, including in R7 cells.
Dotted blue box represents 23% co-expression with Pros in R7, while the dotted white box represents 4% co-expression in another part of the retina.
Additional files
-
Source code 1
Custom Python source code.
- https://doi.org/10.7554/eLife.25301.011
-
Source data 1
Source data for all figures
R8 photoreceptor subtype counts for each scored retina for all genotypes are listed with calculated average percentages of Rh5, Rh6, and Rh5 + Rh6 co-expressing cells. Standard deviation and standard error are also calculated for all groups. Genotype averages with standard error are presented and color coded in columns A-G (blue = Rh5, purple = Rh5 + Rh6 co-expression, red = Rh6).
- https://doi.org/10.7554/eLife.25301.012
-
Transparent reporting form
- https://doi.org/10.7554/eLife.25301.013





