MeCP2 regulates Tet1-catalyzed demethylation, CTCF binding, and learning-dependent alternative splicing of the BDNF gene in Turtle
Figures
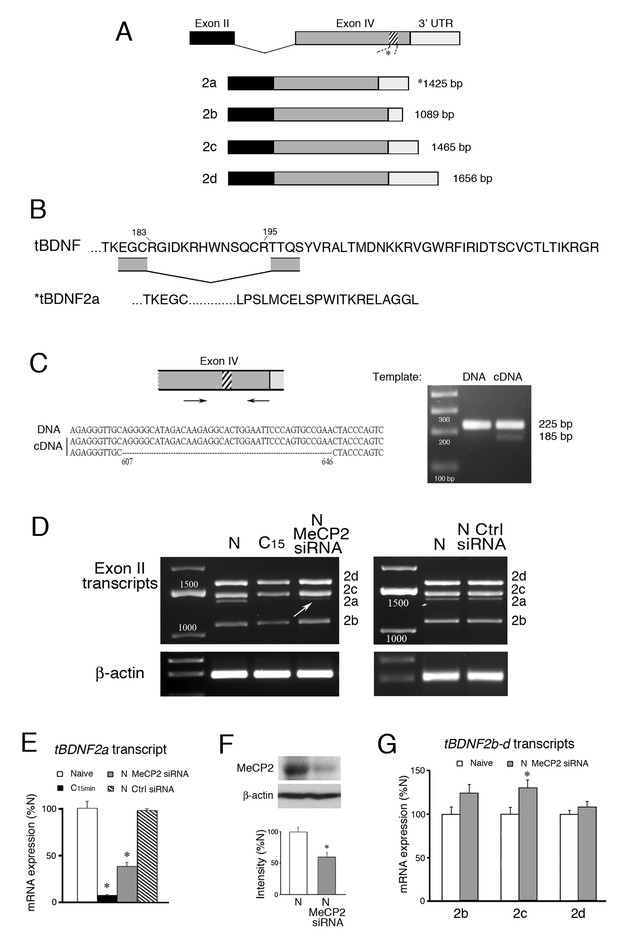
Conditioning-dependent alternative splicing of tBDNF is mediated by MeCP2.
(A) Schematic diagram of tBDNF mRNA transcripts generated from non-coding exon II and the protein coding exon IV. Four transcripts designated tBDNF2a-d are produced in naïve preparations but only the tBDNF2a transcript undergoes an intraexonic splicing event in which 40 bp (13 amino acids) are removed from the distal region of the coding sequence (shown by the hatching at nt 607–646). (B) The amino acid sequence of the distal end of the protein coding sequence is shown for the full-length tBDNF transcripts. The deletion of the 13 amino acids in tBDNF2a (aa 183–195) is also shown which results in a frame shift and alternative C-terminal end with an early stop codon that generates the truncated tBDNF protein. Complete sequences of the tBDNF and tBDNF2a preproBDNF proteins are shown in Ambigapathy et al. (2013). (C) The region of exon IV that undergoes the splicing event was further analyzed using primers flanking the splice site. The PCR products generated from genomic DNA produced a single band at 225 bp while cDNA produced two bands at 225 bp and 185 bp. Sequencing showed that the larger PCR band from cDNA was identical to tBDNF2b-d, while the smaller band was tBDNF2a (accession numbers: KC151267 – KC151270). (D) Four tBDNF exon II transcripts 2a-d are expressed in naïve (N) preparations. After 15 min of conditioning (C15), all transcripts are downregulated but only the 2a transcript is nearly completely suppressed. Application of a MeCP2 siRNA (200 nM, 24 hr) to naïve preparations inhibits tBDNF2a expression (arrow) while a control siRNA (Ctrl siRNA; 200 nM, 24 hr) does not. (E) Semi-quantitative data of tBDNF2a mRNA expression in the different experimental conditions is shown relative to naive. The tBDNF2a transcript is significantly reduced during conditioning and after treatment with MeCP2 siRNA. (F) Western blots confirm that the MeCP2 siRNA significantly inhibits total MeCP2 protein compared to normal naïve. (G) Expression of the remaining exon II transcripts, tBDNF2b-d, is not inhibited by application of MeCP2 siRNA. For this and all figures, p and n values are given in the text.
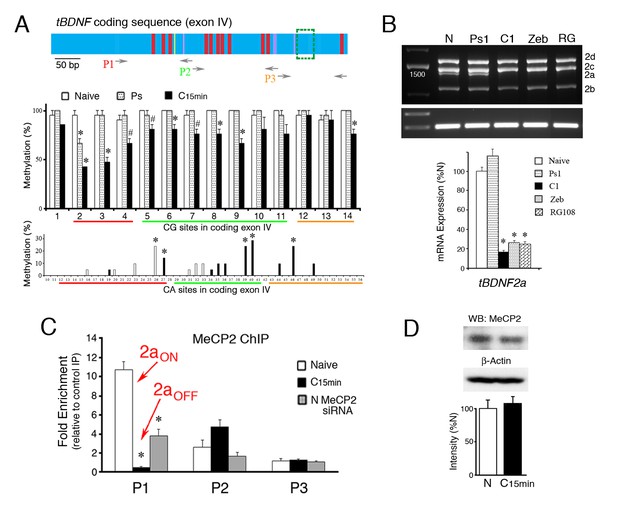
Suppression of tBDNF splicing during conditioning involves demethylation and release of MeCP2 binding.
(A) Schematic illustration of the tBDNF protein coding sequence (upper panel) showing demethylated CG sites during conditioning in red, demethylated CA sites in yellow, and methylated CA sites in purple. The region deleted by splicing is indicted by the green box. The relative coverage of primer pairs P1-P3 used for ChIP-qPCR is indicated. The middle panel shows the methylation status of the tBDNF coding sequence determined by bisulfite sequencing PCR (BSP) in naïve (N), pseudoconditioned (Ps), and conditioned preparations (C15 min). Nearly all CG sites are significantly demethylated after conditioning for 15 min. 3 × 7 clones/group. As shown in the lower panel, specific CA sites also undergo conditioning-related methylation or demethylation. Bars in white are sites that are methylated in naïve and completely demethylated to 0% in conditioning; bars in black are unmethylated in naïve and methylated in conditioning. 3 × 10 clones/group. Red, green, and yellow lines indicate coverage by primer sets P1-P3, respectively. #p<0.05; *p<0.01. (B) Expression of the spliced tBDNF2a transcript is significantly inhibited relative to naïve by the DNMT inhibitors zebularine (Zeb; 100 µM) or RG108 (RG; 200 ng/µl) applied to the bath as shown by the semi-quantitative analysis. Levels of tBDNF2a transcripts from pseudoconditioned (Ps1) or conditioned (C1) preparations for one pairing session are also shown. (C) ChIP-qPCR assays for total MeCP2 protein show that primer set P1 detects MeCP2 tightly bound to tBDNF in naïve preparations during splicing of tBDNF2a (2aON) and is released after conditioning when splicing stops and tBDNF2a is suppressed (2aOFF). Application of the MeCP2 siRNA to naïve preparations results in significantly reduced ChIP signal in the P1 region compared to normal naïve. (D) Western blots confirm that levels of total MeCP2 are not altered during conditioning.
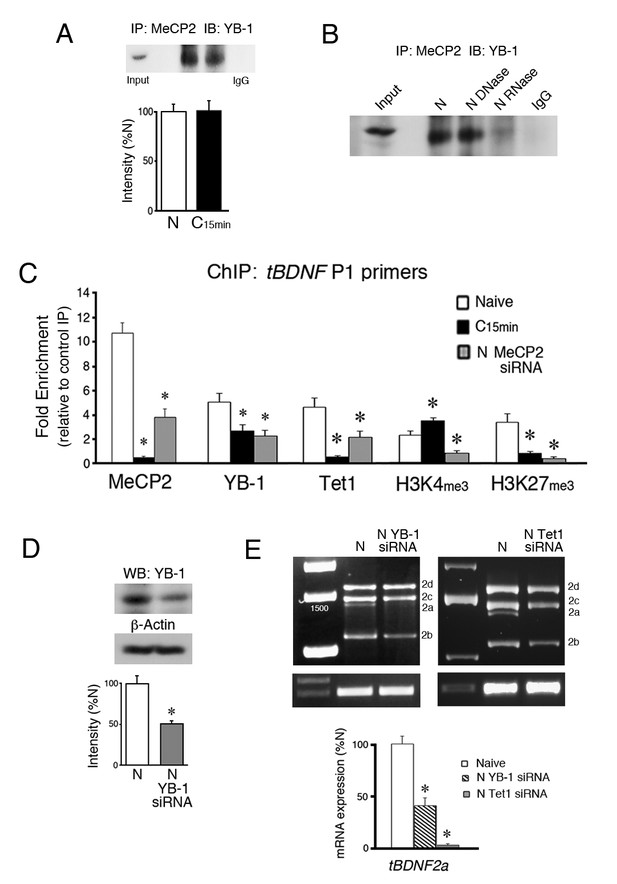
YB-1 and Tet1 partner with MeCP2 during splicing and are inhibited from tBDNF binding by MeCP2 siRNA.
(A) Coimmunoprecipitation of MeCP2 with YB-1 shows a strong interaction in both naïve and conditioned preparations. A control co-IP is omitted because total MeCP2 protein immunoblots at a molecular weight similar to heavy chain IgG. IP, immunoprecipitation; IB, immunoblot. (B) Treatment of naïve brainstem samples with DNase had no effect on coimmunoprecipitation of MeCP2 with YB-1 while RNase treatment inhibited the interaction. (C) ChIP-qPCR assays of tBDNF binding in the region of the coding sequence surveyed by primer set P1. Data are from analysis of tissue samples from naïve preparations, those conditioned for 15 min (C15 min), and naïve treated with MeCP2 siRNA (N MeCP2 siRNA). *, Significant differences from naïve. (D) Western blots show that YB-1 siRNA (200 nM, 24 hr) significantly reduces expression of YB-1 protein to 53% of naïve. (E) The tBDNF2a transcript is significantly inhibited by siRNAs targeting either YB-1 or Tet1 applied to naïve preparations. Data for the Tet1 siRNA (150 nM, 24 hr) are replotted from Ambigapathy et al. (2015).
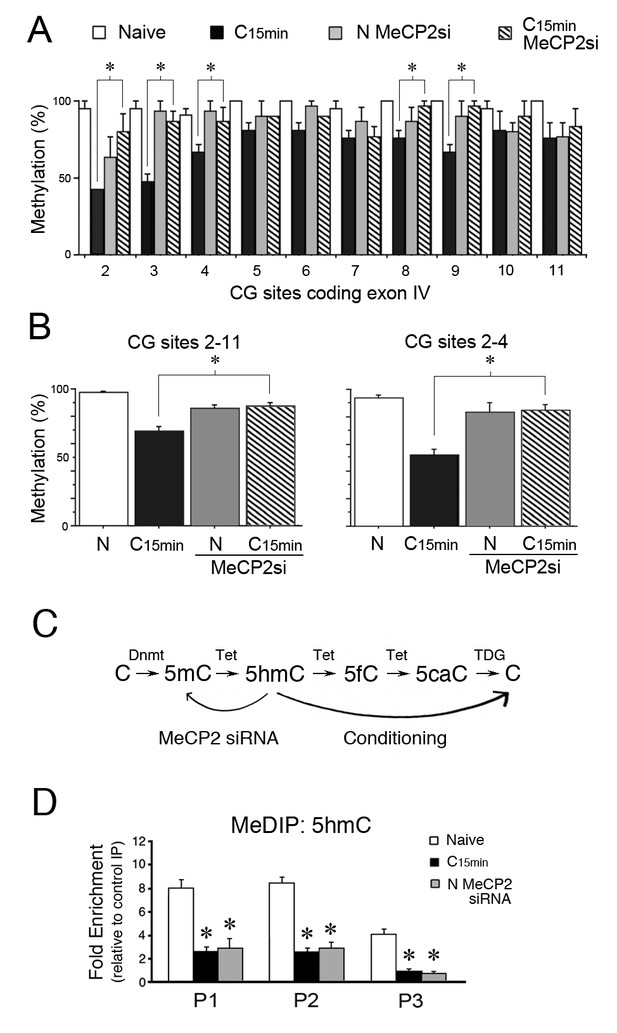
MeCP2 siRNA inhibits demethylation of the tBDNF coding sequence during conditioning.
(A) BSP analysis of the coding sequence in naïve, conditioned preparations, and those treated with MeCP2 siRNA. Specific CG sites demethylated in normal conditioning show significantly greater levels of methylation after conditioning in MeCP2 siRNA. Data from normal naïve and conditioned are from Figure 2 and are shown here for comparison. MeCP2 siRNA data are from 3 × 10 clones/group. *Significant differences (p=0.01) between the C15 min MeCP2 siRNA treated group compared to normal C15 min. (B) Grouped data of CG sites 2–11 (left panel) and sites 2–4 (right panel) show significant attenuation of demethylation in conditioned preparations treated with MeCP2 siRNA compared to normal conditioning. (C) Schematic illustration of the putative stepwise oxidative demethylation pathway performed by Tet proteins and the thymine DNA glycosylase (TDG) and base excision repair system. Normal conditioning drives the process to the right toward unmethylated C, whereas treatment of naïve or conditioned preparations with MeCP2 siRNA that results in a reduction of Tet1 binding to tBDNF shifts the pathway to the left toward 5mC. 5fC, 5-formylcytosine; 5caC, 5-carboxylcytosine. (D) MeDIP assays show high levels of 5hmC content in the tBDNF coding sequence of naïve preparations. These levels are significantly reduced after normal conditioning and in naïve preparations treated with MeCP2 siRNA.
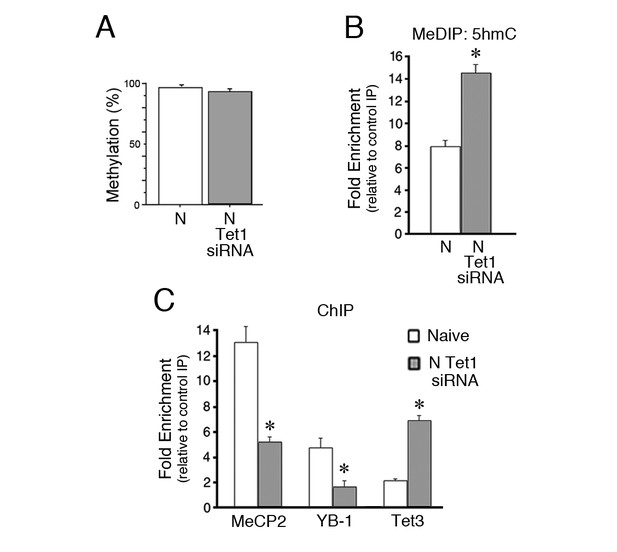
Inhibition of Tet1 by siRNA alters methylation and blocks MeCP2 and YB-1 binding to the coding sequence.
(A) Methylation status of the tBDNF coding sequence (CpGs 2–11) in normal and Tet1 siRNA-treated naïve preparations analyzed by BSP. High levels of methylation were detected for both conditions. Normal naïve data are reshown here from earlier figures; Tet1 siRNA data are from 3 × 10 clones/group. (B) MeDIP assay showing the substantial increase in 5hmC in the P1 primer set region of the coding sequence after treatment of naïve preparations with Tet1 siRNA. Data from normal naïve are reshown from Figure 4D; n = 3 for Tet1 siRNA group. (C) ChIP assays reveal a significant reduction in binding of MeCP2 and YB-1 to the coding sequence at P1 after Tet1 siRNA treatment. In contrast, binding for Tet3 in conditions of Tet1 siRNA was dramatically increased. Data for YB-1 normal naïve are reshown from Figure 3C for comparison.
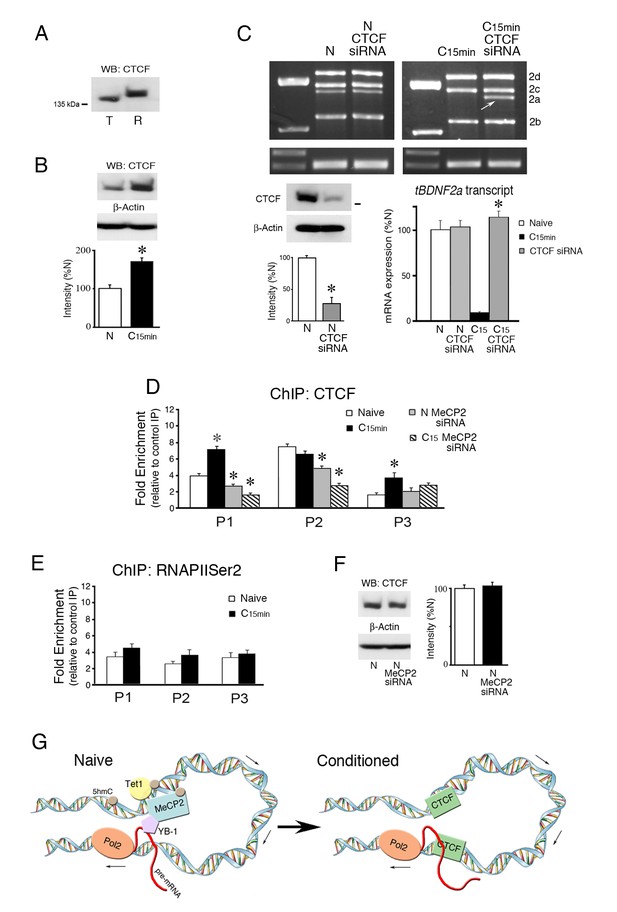
The insulator protein CTCF suppresses splicing of the tBDNF2a transcript during conditioning.
(A) The CTCF antibody recognizes an ~140 kDa protein band in both turtle (T) and rat (R) brain tissue. (B) CTCF protein expression in turtle is significantly upregulated during conditioning. (C) Western blots show that the CTCF siRNA significantly inhibits CTCF protein expression compared to naïve (lower left panel). Expression of exon II transcripts after treatment with CTCF siRNA results in no change in the level of spliced tBDNF2a in treated naïve preparations, as shown by the PCR gels (upper panels) and semi-quantitative data (lower right panel). In conditioned preparations (C15 min) in which tBDNF2a is normally suppressed, CTCF siRNA application results in strong expression of tBDNF2a (arrow) to near normal naïve levels. *p<0.0001, C15 CTCF siRNA vs. C15. (D) ChIP assays of CTCF show that binding to tBDNF is significantly increased after conditioning in regions surveyed by primers P1 and P3. Application of MeCP2 siRNA significantly reduces binding in both naïve (N) and conditioned (C15) preparations in regions P1 and P2 compared to normal naïve. (E) ChIP of RNAPIISer2 binding to the coding exon shows no significant changes during conditioning compared to naïve. (F) Expression of CTCF protein is not affected by application of the MeCP2 siRNA. (G) Model summarizing the molecular events during splicing in naïve and conditioned preparations. The MeCP2 DNA binding region (surveyed by primer set P1) is upstream of the splice site (surveyed by primers P3). We hypothesize they are drawn into close proximity of one another by chromatin looping. In naïve preparations, DNA in the MeCP2 binding region is methylated with high 5hmC content maintained by Tet1 that colocalizes with MeCP2. MeCP2 recruits YB-1 that performs the splicing as the nascent transcript is elongated by RNAPII (Pol2) at the downstream leg of the loop to generate tBDNF2a. Upon conditioning, 5hmC is demethylated to unmodified C and Tet1 dissociates along with MeCP2 and YB-1. This is followed by new occupancy by CTCF that insulates DNA from further splicing thereby suppressing tBDNF2a during conditioning.
Tables
Primers used for BSP analysis of tBDNF coding sequence.
| Target | Orientation | Sequence (5’→ 3’) |
|---|---|---|
| CG 1–4 | For | AAGTGTTAGAGGATTAGGTAGTTTGGTTTATTTAGGT |
| Rev | AAAAACAATAAAAACTCCAAAAACAC | |
| CG 2–11 | For | ATGTTATAGAGGAGTTTTTAGATGAGGA |
| Rev | TACCTCTTATCTATACCCCTACAACCCTCTTT | |
| CG 12–14 | For | AGAGGGTTGTAGGGGTATAGATAAGAGGTA |
| Rev | CGATTCTTAACAACGACAACAAACCACAA |
Primers used for ChIP and MeDIP assays of tBDNF coding sequence.
| Target | Orientation | Sequence (5’→ 3’) |
|---|---|---|
| P1 | For | AGCCTAAGTGGGCCCAACAC |
| Rev | TCCTCAAGCAGAAAGAGCAATG | |
| P2 | For | GATGCTGCAAATATGTCCATGAG |
| Rev | TCAGTTGGCCTTTGGGTACTG | |
| P3 | For | CAAATGCAATCCCAAAGGTTACACAA |
| Rev | TCTTATAAACCGCCAGCCAACT |
Additional files
-
Supplementary file 1
Raw sequence data from splice site PCR and RT-PCR.
Sequence data from PCR products generated from analysis of the splice site shown in Figure 1C. Sequence from the single band produced from genomic DNA is shown in sample 1. Data from cDNA produced two PCR bands whose sequences are shown for the upper band (sample 2) and the lower band (sample 3). ^Indicates the splice site in sample 3.
- https://doi.org/10.7554/eLife.25384.010





