Inter-Fork Strand Annealing causes genomic deletions during the termination of DNA replication
Figures
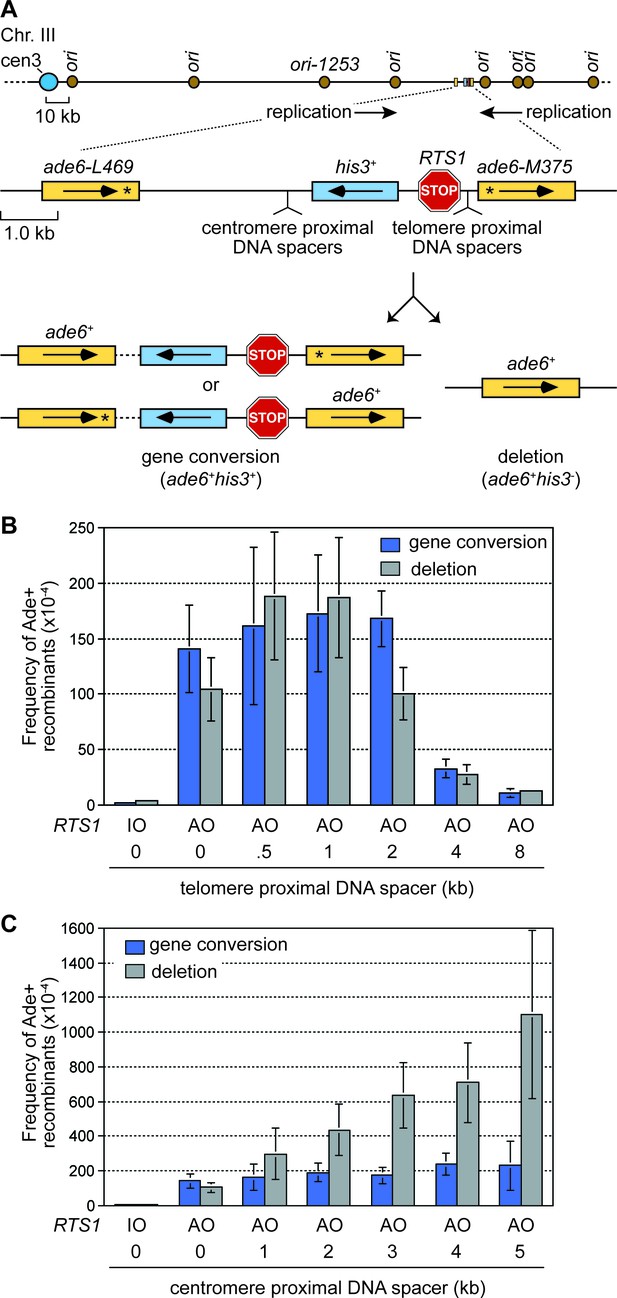
The effect of inter-DNA repeat distance on RF barrier-induced recombination.
(A) Schematic showing the location of RTS1, ade6- direct repeats, sites of DNA spacer insertion, nearby replication origins, and products of inter-repeat recombination. Asterisks indicate the position of point mutations in the ade6 alleles. (B to C) Mean frequency of Ade+ gene conversions and deletions in wild-type cells with RTS1-IO or -AO and different length telomere- and centromere-proximal DNA spacers. Error bars represent 1 SD.
-
Figure 1—source data 1
Effect of inter-repeat distance on the frequency of RTS1-AO-induced direct repeat recombination.
- https://doi.org/10.7554/eLife.25490.003
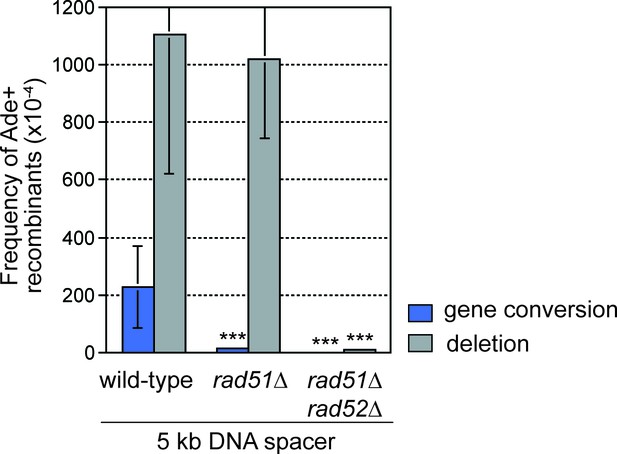
DNA spacer-dependent deletions depend on Rad52 but not Rad51.
Mean frequency of Ade+ gene conversions and deletions in wild-type, rad51∆ and rad51∆ rad52∆ strains containing RTS1-AO with a 5 kb centromere-proximal DNA spacer. Significant fold changes compared to the wild-type strain are indicated (***p<0.001). Error bars represent 1 SD.
-
Figure 2—source data 1
Frequency of RTS1-AO-induced direct repeat recombination in wild-type, rad51∆ and rad51∆ rad52∆ strains with an extra 5 kb DNA spacer between the repeats.
- https://doi.org/10.7554/eLife.25490.005
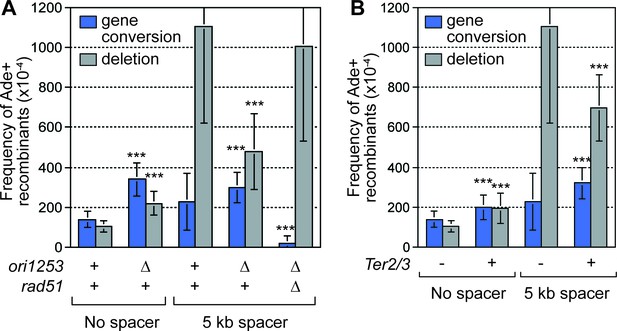
Deletion of the downstream replication origin ori1253, or introduction of a Ter2/3 barrier downstream of RTS1-AO, reduces the frequency of SDDs.
(A) Mean frequency of Ade+ gene conversions and deletions in wild-type and ori1253∆ strains containing RTS1-AO with and without a 5 kb centromere-proximal DNA spacer. The effect of deleting rad51 on the frequency of recombinants in a ori1253∆ strain with a 5 kb centromere-proximal DNA spacer is also shown. (B) Effect of a Ter2/3 barrier downstream of RTS1-AO on the frequency of Ade+ gene conversions and deletions in wild-type strains with and without a 5 kb centromere-proximal DNA spacer. Significant fold changes compared to the equivalent wild-type strain are indicated (***p<0.001). Error bars represent 1 SD.
-
Figure 3—source data 1
Effect of delaying fork convergence on the frequency of RTS1-AO-induced direct repeat recombination with and without an extra 5 kb DNA spacer between the repeats.
- https://doi.org/10.7554/eLife.25490.007
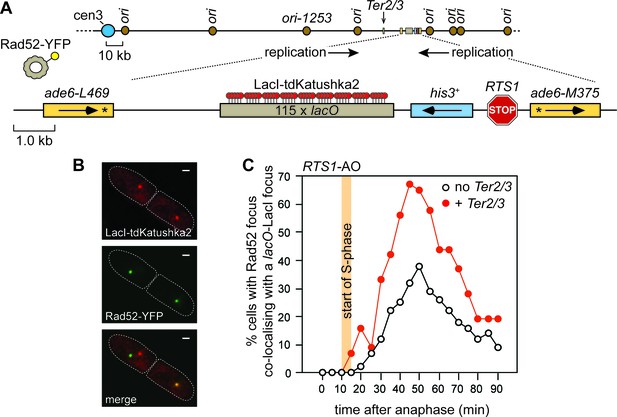
The effect of delaying the incoming RF, with a Ter2/3 barrier, on recruitment of Rad52 to RTS1.
(A) Schematic showing the position of the Ter2/3 replication fork barrier downstream of RTS1-AO on chromosome 3. For the live cell imaging experiments, in B) and C), a lacO array is also inserted downstream of RTS1-AO to enable tracking of the locus, in cells expressing LacI-tdKatushka2, by time-lapse fluorescent microscopy. Rad52 is tagged with yellow fluorescent protein (YFP) and forms nuclear foci that mark sites of recombination activity. Co-localization of a Rad52-YFP focus with a lacO-LacI-tdKatushka2 focus indicates recombination activity at RTS1 (Nguyen et al., 2015). (B) Representative stills taken from a time-lapse movie of a wild-type cell showing a pair of daughter cells (outlined by white dashed lines) each with a Rad52-YFP focus. In the right-hand cell the Rad52-YFP focus co-localizes with a lacO-LacI-tdKatushka2 focus that marks the position of RTS1-AO in the nucleus. The scale bar represents 1 µm. (C) The effect of Ter2/3 on the percentage of cells with a Rad52-YFP focus co-localizing with a lacO-LacI-tdKatushka2 focus in the first 90 min period post-anaphase. S-phase starts 10–15 min after anaphase (Nguyen et al., 2015). Delaying the incoming fork, by inserting a Ter2/3 barrier downstream of RTS1-AO, provides more time for Rad52 recruitment to the RF blocked at RTS1, as evidenced by the greater percentage of cells with a Rad52-YFP focus co-localizing with a lacO-LacI-tdKatushka2 focus. This result indicates that RF convergence and termination at RTS1-AO prevents Rad52 recruitment, and agrees with the finding of a previous experiment where incoming replication was delayed by deleting ori1253 (Nguyen et al., 2015). The strain with no Ter2/3 is MCW6556 (n = 123) and the strain with Ter2/3 is MCW7304 (n = 44).
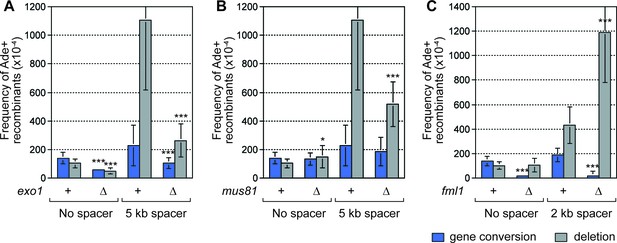
SDDs depend on Exo1 and Mus81, but are suppressed by Fml1.
(A–C) Mean frequency of Ade+ gene conversions and deletions in wild-type and mutant strains containing RTS1-AO with and without a centromere-proximal DNA spacer as indicated. Significant fold changes compared to the equivalent wild-type strain are indicated (*p<0.05, **p<0.01, ***p<0.001). Error bars represent 1 SD.
-
Figure 4—source data 1
Frequency of RTS1-AO-induced direct repeat recombination in wild-type, exo1∆, mus81∆ and fml1∆ strains with and without an extra DNA spacer between the repeats.
- https://doi.org/10.7554/eLife.25490.010
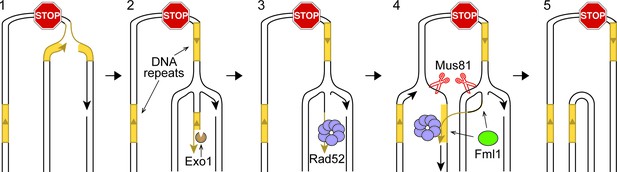
IFSA model.
See main text for details.
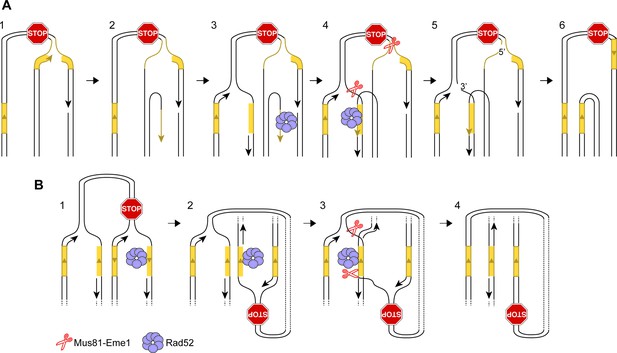
Alternative models for IFSA.
(A) IFSA initiated from an unwound nascent leading strand. Step 1: RF collapses following stalling at a RFB. Step 2: A DNA helicase unwinds the nascent leading strand at a repetitive DNA sequence. Step 3: The unwound nascent leading strand is bound by RPA followed by Rad52. Step 4: Rad52 anneals the nascent leading strand to a complementary repeat sequence exposed in the lagging strand gap of the incoming RF. Steps 4–5: Mus81-Eme1 cleaves both RFs generating one full-length sister chromatid, containing single-strand gaps, and one shortened sister chromatid with a 3’ and 5’ flap. Note that the 5’ flap can anneal to the nascent strand causing the formation of a longer 3’ flap. Step 6: Gap filling, flap removal and ligation of strand nicks, results in one intact full-length sister chromatid and one lacking a DNA repeat and the inter-repeat region. (B) IFSA by annealing between lagging strand gaps. Step 1: Following RF collapse at the RFB, Exo1 degrades the lagging nascent strand exposing ssDNA at a repetitive sequence, which is bound by RPA and Rad52. Step 2: As the incoming RF converges on the collapsed fork, it comes into close proximity with the ssDNA bound by Rad52. Step 3: Rad52 anneals its bound ssDNA to the complementary ssDNA exposed in the lagging strand gap of the incoming fork. Steps 3–4: Mus81-Eme1 cleaves the DNA junctions at either end of the annealed region to resolve the sister chromatids into full-length and shortened forms. As with the other versions of the IFSA model, it is expected that gap filling, flap removal and ligation of strand nicks will be needed to convert the products of Mus81 cleavage into intact sister chromatids. DNA repeats are marked in yellow.
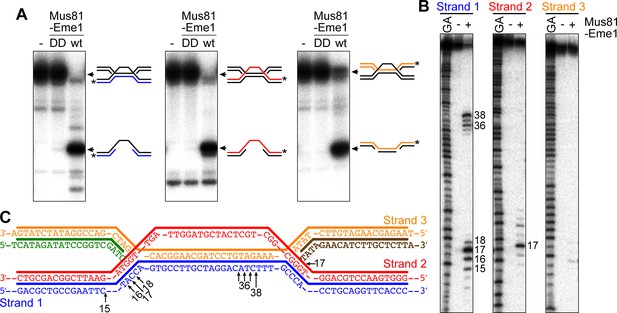
Cleavage of a model IFSA junction by Mus81-Eme1.
(A) Native gel analysis of Mus81-Eme1 cleavage reactions with wild-type (wt) and catalytically inactive (DD) enzyme. The schematics show the IFSA junction and cleavage products. The asterisks indicate the position of the 5′-end 32P label. (B) Denaturing gel analysis of Mus81-Eme1 cleavage reactions to map the sites of strand cleavage by reference to a G and A Maxam-Gilbert sequencing ladder of the relevant labelled strand. (C) Schematic showing the sites of strand cleavage (arrows).
Additional files
-
Supplementary file 1
Schizosaccharomyces pombe strains.
- https://doi.org/10.7554/eLife.25490.014
-
Supplementary file 2
Oligonucleotides.
- https://doi.org/10.7554/eLife.25490.015





