The Anopheles gambiae 2La chromosome inversion is associated with susceptibility to Plasmodium falciparum in Africa
Figures
African study sites used in study of the 2La inversion.
Samples of Anopheles gambiae complex from sites in Kenya (dark green), Republic of Guinea (Guinea-Conakry, orange) and Burkina Faso (blue) were used to assess the association between the 2La inversion and susceptibility to Plasmodium falciparum infection presented in Figure 2. Samples collected from Kenya, Guinea-Conakry, Cameroon (yellow), Uganda (light green), Burkina Faso, and Guinea Bissau (red) were also used to assess the phylogenetic relationship of 2La inversion alleles, presented in Figure 6.
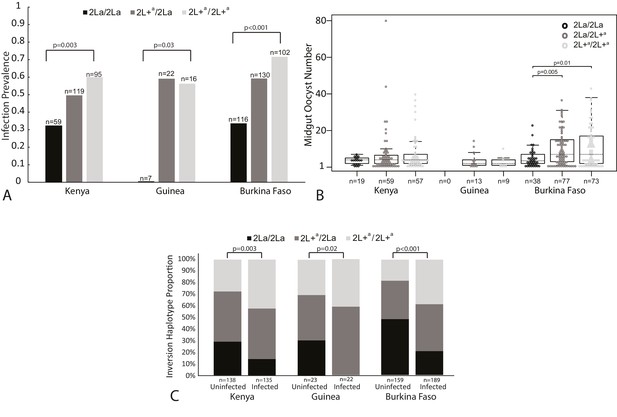
Plasmodium falciparum infection is associated with the 2La inversion genotype in the Anopheles gambiae complex across Africa.
(A) Oocyst infection prevalence. Sampled mosquitoes were dissected to count midgut oocysts and were molecularly genotyped for the 2La inversion. Differences in oocyst infection prevalence between all three inversion genotypes are indicated by vertical bars, and statistical difference between inversion homozygotes, 2La/2La and 2L+a/2+a, at each geographic sampling site are indicated by p-values. Pairwise statistical comparisons between all inversion genotypes are presented in Figure 2—figure supplement 1. Oocyst infections were measured in wild-caught adult mosquitoes after uncontrolled exposure to bloodmeals and parasites in nature (Kenya and Guinea-Conakry), or in adults grown from wild larvae, then membrane-fed on blood from natural gametocyte carriers (Burkina Faso). In Kenya, mosquitoes were held for 7 days before dissection, whereas in Guinea-Conakry, mosquitoes were dissected upon capture. Association was measured in test sets including all infected mosquitoes and an equal number of random non-infected mosquitoes from the same collection, to normalize average oocyst infection prevalence to ≥50% for each site (see Materials and methods). This analysis provides the statistical power to detect both increase and decrease of in prevalence, and normalizes statistical power across all sites. The Burkina Faso collection was comprised of multiple taxa, which are presented and analyzed separately in Figure 2—figure supplement 2. (B) Oocyst infection intensity. The numbers of midgut oocysts 7–8 days after infection are shown for all individuals with ≥1 midgut oocyst. Statistical analysis by non-parametric Wilcoxon-Mann-Whitney tests indicates that within the Burkina Faso samples, the 2La/2La mosquitoes had significantly lower parasite loads as compared to 2La/2L+a or 2L+a/2L+a. A similar trend is evident though non-significant in the Kenya samples (all p values shown in Figure 2—figure supplement 1). Boxplots delineate the first and third quartiles, median is indicated by the dashed line within the box and error bars are 1.5 time the interquartile range. (C) 2La genotype frequency as a function of infection outcome. At each sampling site, 2La/2La homozygotes are more prevalent in the uninfected group, whereas 2L+a/2L+a homozygotes are more prevalent in the infected group. 2La inversion frequencies are significantly different between the uninfected and infected samples at each of the three sampling sites.
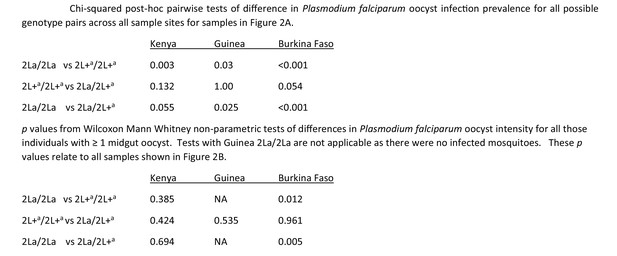
p-Values for all Chi-Square post-hoc comparisons.
Significance values for all pairwise comparisons from the analysis of infection prevalence (Figure 2A) and oocyst intensity (Figure 2B).
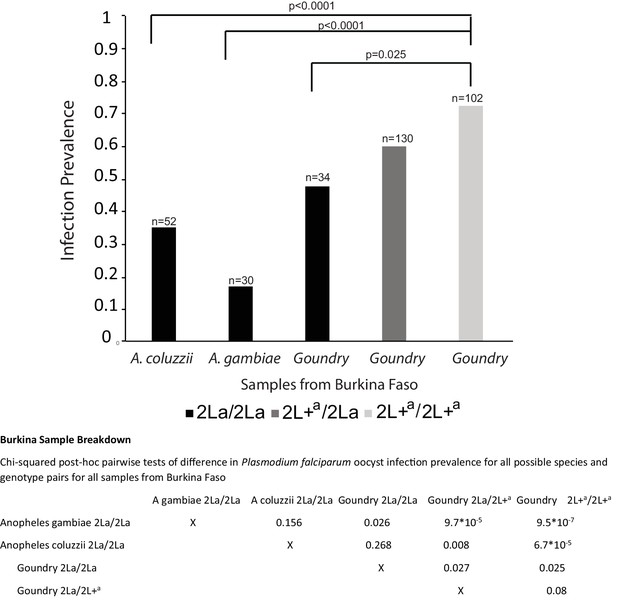
Taxon breakdown and infection analysis of samples from Burkina Faso.
Data for samples from Burkina Faso separated by the mosquito taxa present in the Burkina Faso collections. The 2L+a/2L+a mosquitoes display significantly greater infection prevalence than 2La/2La homozygotes in all comparisons, either within the Goundry form or compared to A. gambiae and A. coluzzii, which do not segregate for the 2La inversion at this geographic site. Pairwise p-values are shown below the figure.
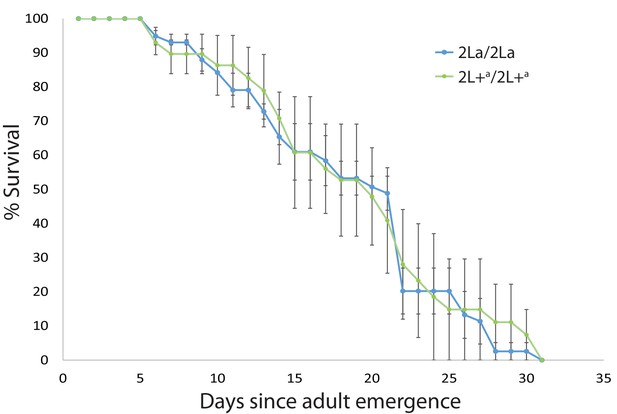
2La inversion homozygotes display equivalent longevity under semi-field conditions.
Wild A. gambiae larvae were collected in western Kenya, and the emerged adults were maintained in outdoor cages, exposed to ambient conditions including environmental microbes and pathogens. Dead mosquitoes were collected daily and genotyped for the 2La inversion. Plot indicates average longevity with standard error for 2La/2La and 2L+a/2L+a homozygotes in three replicate experiments (median longevity 19.3 d). The absence of survival difference between 2La inversion homozygotes exposed to natural microbes and pathogens suggests that the higher malaria infection levels of 2L+a/2L+a mosquitoes is not a consequence of general immune deficiency.
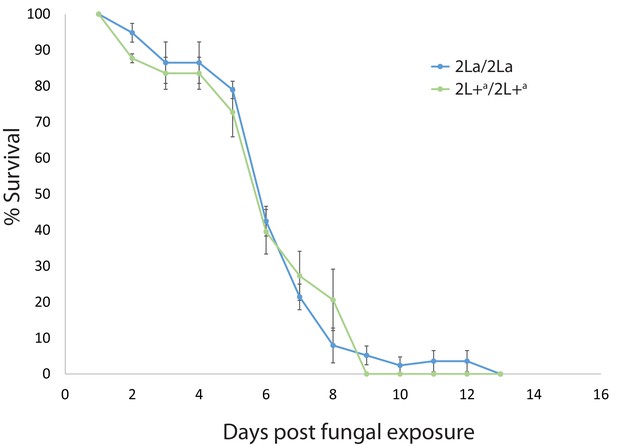
2La inversion homozygotes are equally susceptible to pathogenic fungus.
As a direct test of general immune competence, adult A. gambiae grown from wild larvae in western Kenya were exposed to spores of the insect fungal pathogen, Metarhizium anisopliae strain ICIPE30. Fungus-exposed mosquitoes were maintained in outdoor cages and 2La genotypes were assayed for longevity. Plot indicates average longevity with standard error for 2La/2La and 2L+a/2L+a homozygotes in three replicate experiments. Fungal exposure decreases longevity as compared to non-exposed controls (median longevity 6 days, compare with 19.3 days in Figure 3), but rates of fungal killing were not different between inversion homozygotes. The 2L+a/2L+a mosquitoes are immune competent against Metarhizium but significantly more susceptible to malaria infection, indicating a relatively specific mechanism of susceptibility to malaria, rather than nonspecific low immune competence.
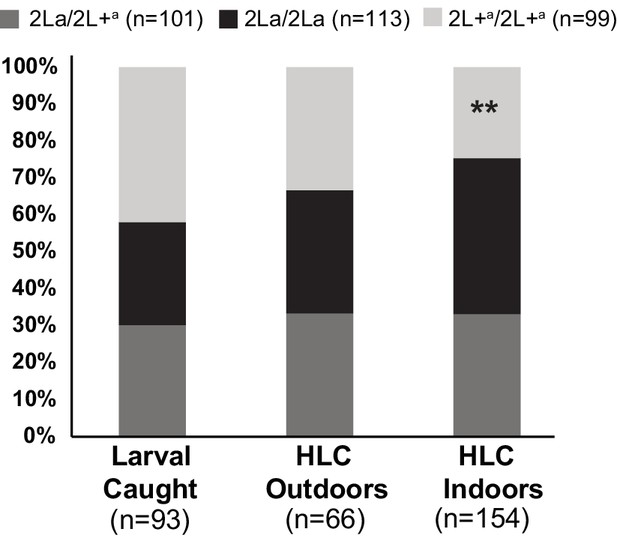
Individuals carrying the 2L+a inversion allele are less likely to be captured indoors.
Adult mosquitoes were sampled in Koraboh and Koundara, Guinea-Conakry by human landing capture (HLC) inside houses or outdoors (≥10 m from the nearest house), and larvae were also collected in the same villages. Of the 2L+a/2L+a homozygotes captured in both villages, a significantly smaller fraction was captured by indoor HLC as compared to the fraction of 2La/2La homozygotes captured by indoor HLC (p=0.008, double asterisk). In addition, there was a significant deficit of 2L+a/2L+a among all captured adults (indoor HLC + outdoor HLC) relative to the expectation from their overall population representation in larval site frequencies, as compared to 2La/2La adults relative to their larval frequencies (p=0.02). The same tendencies are observed for the two individual sites (Figure 5—figure supplement 1).
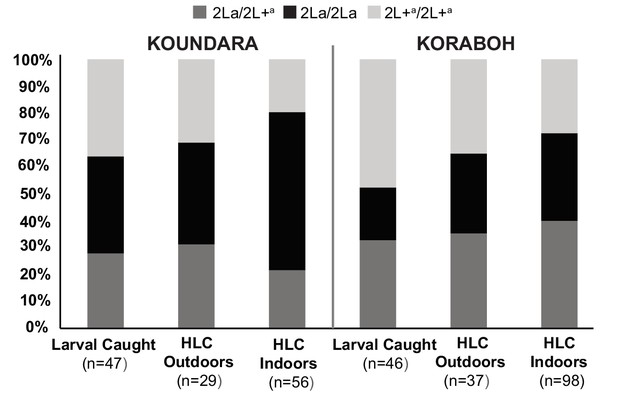
Spatial partitioning of 2L+a carriers is reproduced at individual Guinea-Conakry study sites.
When the Koraboh and Koundara sites were analyzed individually, adult mosquito HLC results display the same tendencies of 2L+a/2L+a deficit indoors, and inefficient human landing capture of 2L+a/2L+a adults overall (Fisher combined p=0.02; individual p-values, Koundara p=0.03, Koraboh p=0.08).
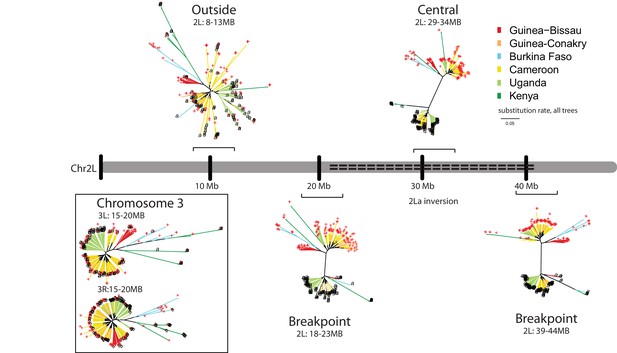
The 2La inversion has a monophyletic origin throughout Africa.
Phylogenetic trees were constructed for genomic windows of 5 megabases (Mb) each extracted from whole-genome sequences of 103 2La/2La and 107 2L+a/2L+a wild A. gambiae mosquitoes, obtained with permission from the Anopheles gambiae 1000 (Ag1000) Genomes Consortium (Miles et al., 2016), and whole-genome sequences of 1 2La/2La and 11 2L+a/2L+a wild Burkina Faso A. gambiae Goundry form mosquitoes (Crawford et al., 2016). Branch color indicates the country origin of the sample (see country key). The terminal label at the end of each branch indicates genotype of the 2La inversion (black ‘a’ for 2La/2La, and red ‘+' for 2L+a/2L+a). The 2La inversion genotypes of Ag1000 samples were determined informatically (Figure 6—figure supplement 1), and Goundry form samples were genotyped by molecular diagnostic assay. The chromosome map depicts the left arm of chromosome 2 (Chr2L), with the centromere to the left, and the dashed line indicates the extent of the 2La inversion. Brackets indicate the genomic windows at three distinct positions relative to the 2La inversion: (i) Spanning the proximal and distal inversion breakpoints (respectively, 2L:18–23 Mb and 2L:39–44 Mb), (ii) in the central region of the inversion (2L: 29–34 Mb), and controls outside the 2La inversion on the same chromosome (2L:8–13 Mb) or on Chromosome 3 (3L:15–20 Mb and 3L:15–20 Mb). Breakpoint and inversion-central trees display a subdivision between 2La inversion genotypes, regardless of the geographic origin of the samples, while trees outside the inversion cluster by geography and not inversion genotype. Branch length scale under the key indicates nucleotide substitutions per site and applies to all six trees.
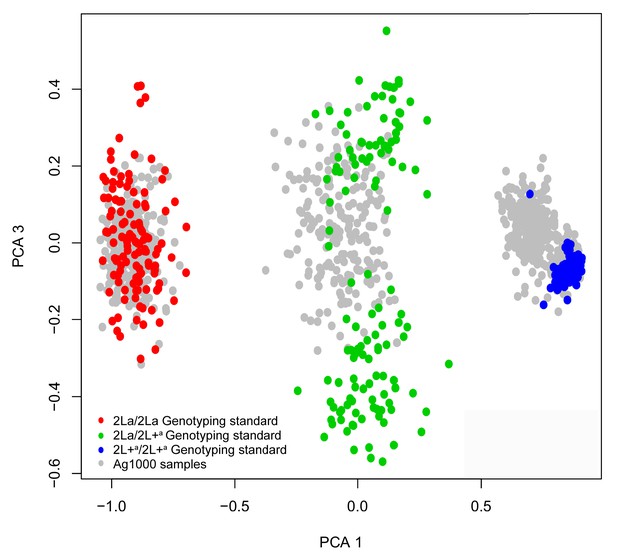
Principal components analysis (PCA) for 2La genotype assignment of whole-genome sequenced samples.
The 2La inversion genotypes of Ag1000 samples were called informatically by supervised clustering, anchored by individuals with independently determined 2La genotypes. Chromosome 2L SNPs of Ag1000 samples (gray points) were clustered by PCA along with the corresponding SNPs of internal genotyping standards with independently determined 2La inversion genotypes (red, green and blue points, see key). The internal genotyping standards were used to paint the Ag1000 sample clusters with genotype calls.
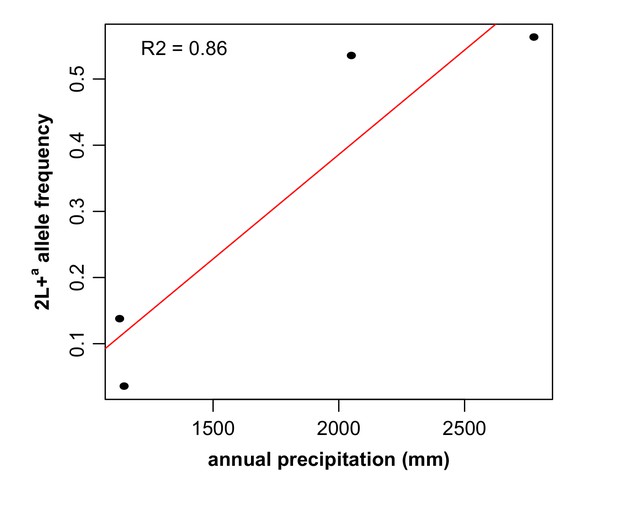
The frequency of the 2La inversion is correlated with ecology.
Frequency of the 2L+a allele was plotted as a function of annual rainfall across an ~350 km ecological transect from arid savanna in southern Mali to deep forest in southern Guinea-Conakry, and displayed a positive correlation (r2 = 0.86, sample sizes from lowest to highest 2L+a frequency, respectively, are n = 190, 191, 365, and 259). Because the samples used in this analysis included individuals from two taxa, A. gambiae and A. coluzzii, an additional analysis of ecological correlation was conducted for each taxon, shown in Figure 7—figure supplement 1.
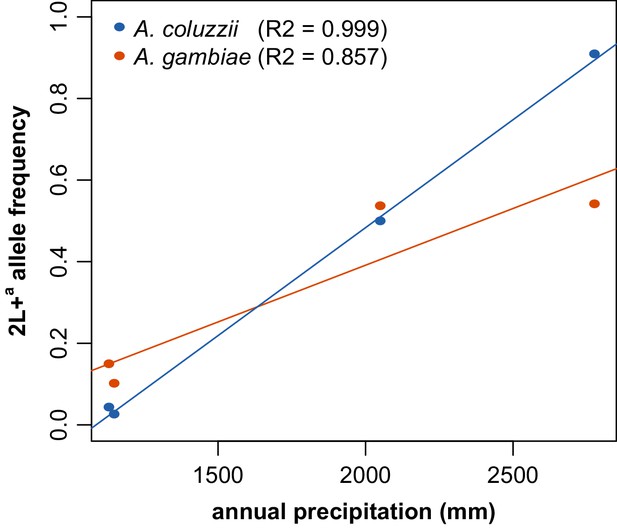
Correlation of 2L+a frequency with annual rainfall is reproduced in individual mosquito taxa.
When the two sister taxa present in Figure 7, A. gambiae and A. coluzzii, were deconvoluted, each still displayed high positive correlation of 2L+a frequency with annual rainfall.





