Inter-dependent apical microtubule and actin dynamics orchestrate centrosome retention and neuronal delamination
Figures
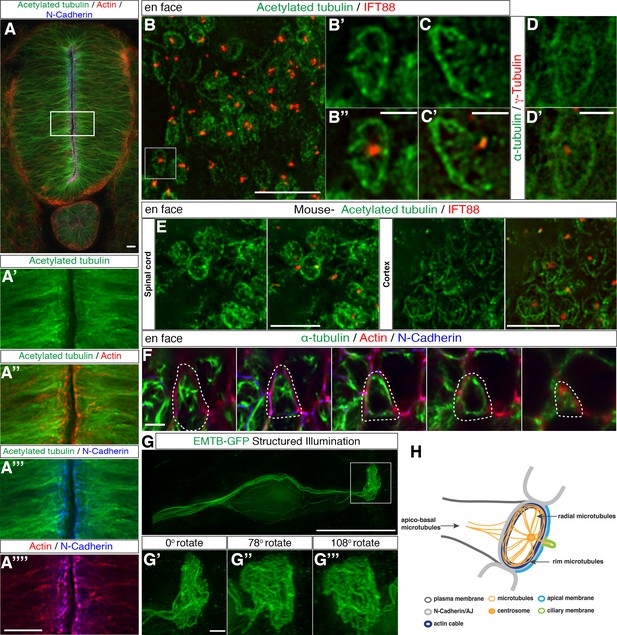
Characterisation of the sub-apical microtubule architecture.
(A) Representative image of a 3-day-old chick embryo neural tube stained with acetylated α-tubulin, phalloidin and N-Cadherin. (A’–A’’’’) Magnification of the boxed region in (A). (B) En face imaging of neuroepithelial end-feet with acetylated α-tubulin and IFT88. (B’–B’’) Magnification of boxed region in (B). (C–C’) Another example as in (B’). (D–D’) End-foot stained with α-tubulin and γ-tubulin. (E) En face imaging of E12.5 mouse embryo spinal cord and cortex stained with acetylated α-tubulin and IFT88. (F) Stills of a neuroepithelial cell (dotted lines show cell outline) en face imaging from apical to more basal (left to right). Tissue explant stained for α-tubulin, N-Cadherin and phalloidin. (G) Neural progenitor cell expressing EMTB-GFP (and nuclear localised GFP from pCIG-Neurog2) imaged with SIM. The boxed region was magnified in (G’–G’’’). Three different angles off the boxed region in G generated by 3D reconstruction. (H) Diagram of microtubule organization at the apical end-feet and relationship with the acto-myosin ring and the AJs. For all figures, images were captured by wide-field microscopy, unless otherwise stated. Scale bars, (A) (B) (E) (G) (A’–A’’’’) 10 μm, (B’–B’’) (C–C’) (D–D’) (F) (G’–G’’’) 2 μm.
-
Figure 1—source data 1
Actin-tubulin co-alignment at the apical adhesion belt level.
- https://doi.org/10.7554/eLife.26215.005
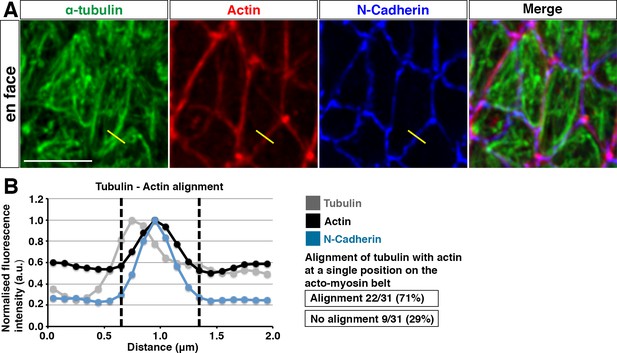
Actin-tubulin co-alignment at the adhesion belt level.
(A) Representative en face image of apical end-feet stained with α-tubulin, phalloidin and N-Cadherin. The yellow line is a representative region to measure fluorescence intensity at a single point on the adhesion belt. (B) Line graphs of normalised fluorescence intensity across the adhesion belt at a single position as in (A). Dashed lines indicate the AJs as defined by N-Cadherin. Scale bar, 5 μm.
Apico-basal Z-stack series across the apical microtubules and sub-apical actin cable and N-Cadherin based adherens junctions; this video is related to Figure 1F.
https://doi.org/10.7554/eLife.26215.0063D structured illumination reconstruction of EMTB-GFP mis-expression at the neuroepithelial cell apical end-foot; this video is related to Figure 1G–G’’’.
The 3D model is rotated 360° on the Y-axis every 3 degrees.
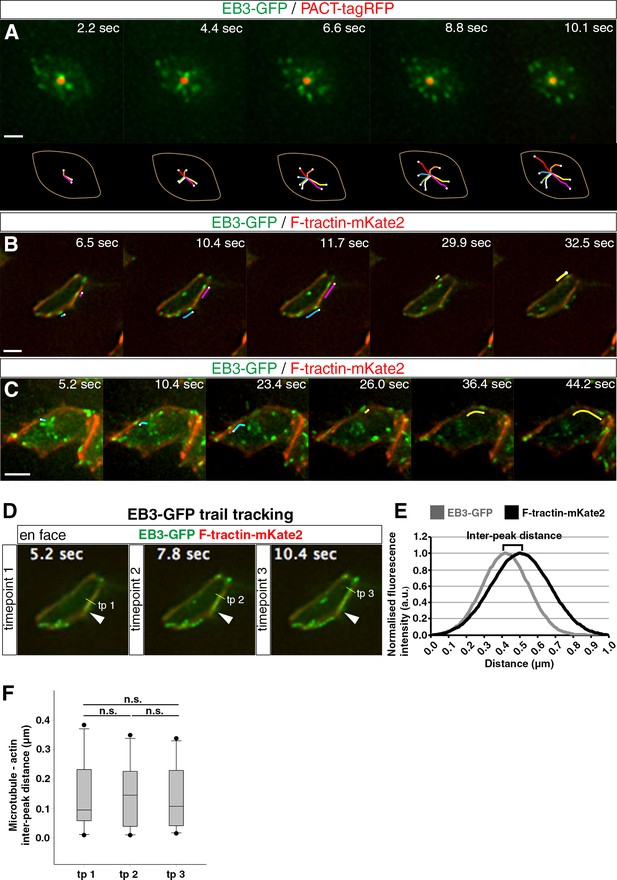
Microtubule dynamics at the apical end-foot and alignment with the actin belt.
(A) Microtubule nucleation from the centrosome. The apical end-foot outline and tracking of EB3-GFP comets over time are shown below. (B) Movement of polymerising microtubules along the actin cable. Lines track movement of two EB3-GFP comets. (C) Microtubules nucleated from the centrosome bend and travel along the actin cable. Lines follow the movement of two EB3-GFP comets. (D) Trail tracking of EB3-GFP comets over time along the F-tractin-mKate2 belt. Three timepoints are shown. The arrowhead represents the starting point of EB3-GFP comet movement. The yellow line shows its position at different timepoints and the method for the measurement of fluorescence intensity at that particular point for both channels. (E) Example of fitted Guassian curves for the calculation of inter-peak distance between the two channels. For the purpose of this example, both fitted fluorescence intensity calculations were normalised from 0 to 1. (F) Box-plots of the microtubule (EB3-GFP)- actin (F-tractin-mKate2) inter-peak distance over time (paired t-test: tp 1 vs tp 2, p=0.84; tp 2 vs tp 3, p=0.72; tp 1 vs tp 3, p=0.96). Scale bars, (A) (B) (C) 2 μm.
-
Figure 2—source data 1
EB3-GFP_F-tractin-mKate2 inter-peak distance.
- https://doi.org/10.7554/eLife.26215.009
Microtubule nucleation from the apical centrosome; this video is related to Figure 2A.
Mis-expressed PACT-TagRFP labels the centrosome and EB3-GFP the growing tips of polymerising microtubules. Maximum intensity projections of deconvolved image sequences are used. Images acquired every 2.2 s.
Microtubule movement along the actin cable; this video is related to Figure 2B.
Mis-expressed F-tractin-mKate2 labels the actin cable and EB3-GFP the growing tips of polymerising microtubules. Maximum intensity projections of deconvolved image sequences are used. Images acquired every 2.6 s.
Trail tracking of EB3-GFP comets; this video is related to Figure 2—video 2 and Figure 2D.
As in Figure 2—video 2. The whole imaging area is represented to the left. To the right a single end foot is selected and magnified, with actin and EB3 channels (top) and only GFP (bottom) Images acquired every 2.6 s.
Microtubule bending at the actin cable; this video is related to Figure 2C.
Mis-expressed F-tractin-mKate2 labels the actin cable and EB3-GFP the growing tips of polymerising microtubules. Maximum intensity projections of deconvolved image sequences are used. Images acquired every 2.6 s.
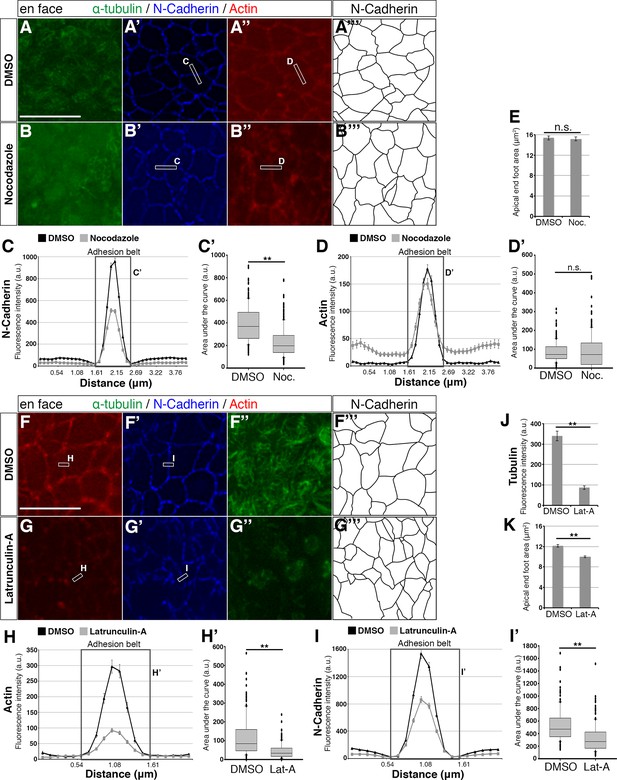
Effects of small molecule treatments on the adhesion belt and microtubules.
(A – A’’, B – B’’, F – F’’, G – G’’) En face imaging of apical end-feet following treatment of chick embryo neural tube explants with Nocodazole (Noc) or Latruncuin-A (Lat-A). Boxed areas indicate how a line is drawn across the adhesion belt for measurement of fluorescence intensity. Letters next to the boxes refer to the corresponding line graphs. (C, D, H, I) Line graphs of normalised fluorescence intensity across the adhesion belt. For Nocodazole a distance of 4 μm and for Latrunculin-A 2 μm was measured. Boxed area represents the adhesion belt and the letter refers to the box plot quantifications from that area. Error bars = SEM. (C’, D’, H’, I’) Box plots of the area under the curve (adhesion belt) from the line graphs. The median value, as well as the upper and lower quartiles are represented. T-test, (C’) p<0.0001 (DMSO [Nocodazole control]: 210 measurements, 6 explants in 3 experiments; Nocodazole: 270 measurements, 8 explants in 3 experiments), (D’) p=0.51 (DMSO [Nocodazole control]: 180 measurements, 6 explants in 3 experiments; Nocodazole: 244 measurements, 8 explants in 3 experiments), (H’) p<0.0001 (DMSO [Latrunculin-A control]: 140 measurements, 5 explants in 2 experiments; Latrunculin-A: 213 measurements, 7 explants in 3 experiments) and (I’) p<0.0001 (DMSO [Latrunculin-A control]: 140 measurements, 5 explants in 2 experiments; Latrunculin-A: 213 measurements, 7 explants in 3 experiments). When the entire curve is considered in (D), the area of the Nocodazole treatment is statistically larger than that of the DMSO treatment, p<0.0001. (E, K) End-foot area measurements for DMSO and small molecule treatments, as outlined by the N-Cadherin staining (A’’’, B’’’, F’’’, G’’’). T-test, (E) p=0.73 (DMSO [Nocodazole control]: 276 measurements in 3 experiments; Nocodazole: 304 measurements in 3 experiments) and (K) p<0.0001 (DMSO [Latrunculin-A control]: 222 measurements in two experiments; Latrunculin-A: 334 measurements in 3 experiments). Error bars = SEM. (J) Normalised tubulin fluorescence following DMSO or Latrunculin-A treatment. T-test, p<0.0001 (DMSO: 110 measurements in 2 experiments; Latrunculin-A: 205 measurements in 3 experiments). Error bars = SEM, scale bars, 10 μm.
-
Figure 3—source data 1
Nocodazole vs DMSO control.
- https://doi.org/10.7554/eLife.26215.015
-
Figure 3—source data 2
Latrunculin-A vs DMSO control.
- https://doi.org/10.7554/eLife.26215.016
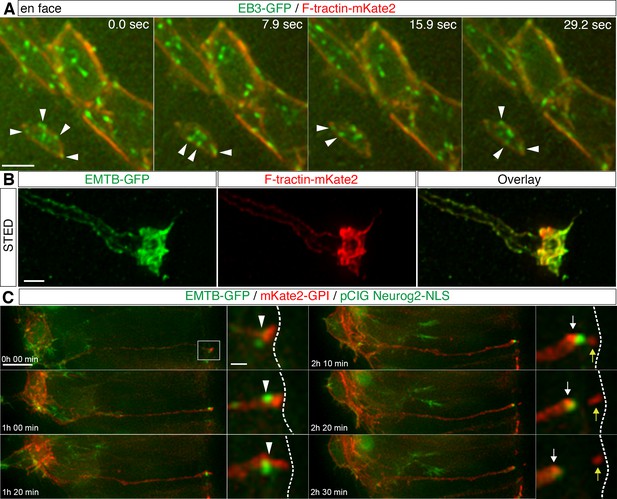
Apical cytoskeletal changes in delaminating cells.
(A) In cells with small apical end-feet, EB3-GFP comets still radiate towards and become closely associated with the actin cable (white arrowheads). (B) STED image of a differentiating neuron end-foot mis-expressing EMTG-GFP (green) and F-tractin-mKate2 (red). (C) Time-lapse sequence of microtubule dynamics during apical abscission. Embryo neural tubes were electroporated with EMTB-GFP (green), pCIG-Neurog2 (nuclear, green) and mKate2-GPI (red). Abscission site (white arrowheads), withdrawing apical process (white arrows), abscised particle (yellow arrows) and apical side (white dashed line). Scale bars, (A) 2 μm, (B) 1 μm, (C) 10 μm, enlarged regions, 2 μm.
Time-lapse sequence of microtubule dynamics in cells with apical end-feet of reduced area; this video is related to Figure 4A.
Mis-expressed F-tractin-mKate2 labels the actin cable and EB3-GFP the growing tips of polymerising microtubules. Maximum intensity projections of deconvolved image sequences are used. Images acquired every 2.6 s.
STED 3D reconstruction of apical end-foot of cell progressing through apical abscission; this video is related to Figure 4B.
Actin is labelled with F-tractin-mKate2 (red) and microtubules are labelled with EMTB-GFP (green). The 3D model is rotated 360o in X- and Y-axes.
Time-lapse sequence of microtubule dynamics during apical abscission; this video is related to Figure 4C.
Mis-expressed Neurog2 (pCIG-Neurog2) labels the nucleus (green). Microtubules are visualized with EMTB-GFP (green) and cell membrane with mKate2-GPI (red). Maximum intensity projections of deconvolved image sequences are used. Images acquired every 10 min.
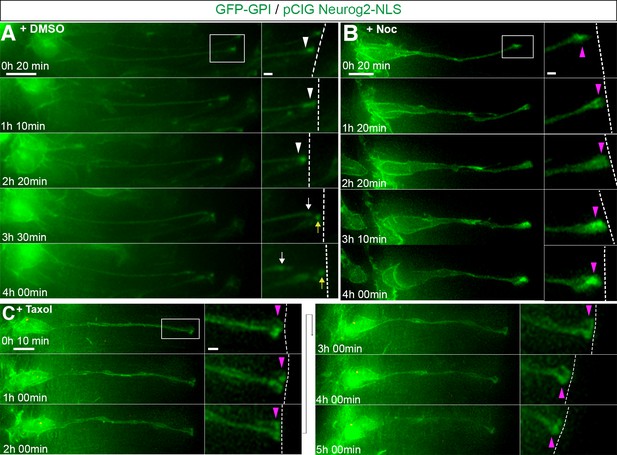
Apical abscission depends on dynamic microtubules.
(A) Time-lapse sequence of cell imaged in medium containing DMSO vehicle control undergoing apical abscission. (B) Time-lapse sequence of cell imaged in medium containing nocodazole. (C) Time-lapse sequence of cell imaged in medium containing taxol. Embryo neural tubes were electroporated with GFP-GPI (cell membrane, green) and pCIG-Neurog2 (nucleus, green). Here and Figure 5—figure supplement 5 : Apical end process (purple arrowhead), abscission site (white arrowheads), withdrawing apical process (white arrows), abscised particle (yellow arrows) and apical side (white dashed line). Scale bars: 10 μm; enlarged region, 2 μm.
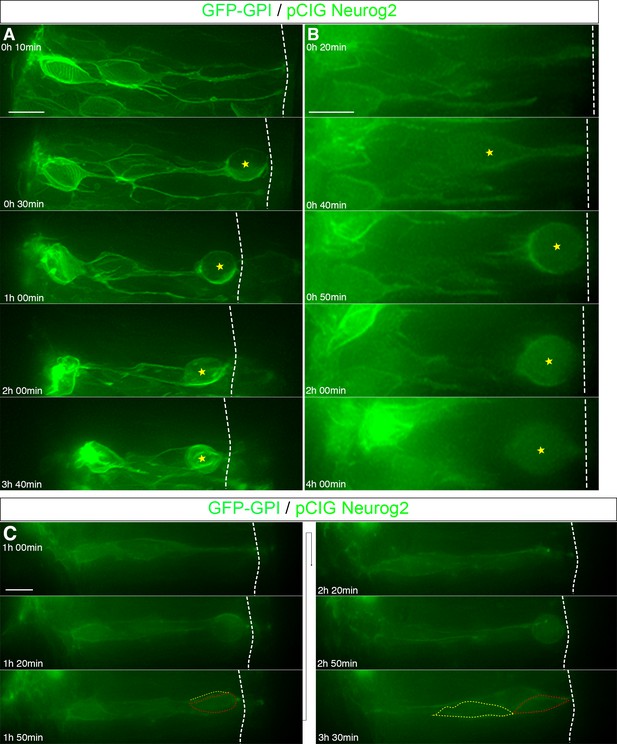
Assessment of nocodazole treatment efficacy in neural tube slices.
(A–B) Two examples of mitotic arrest following nocodazole treatment. Arrested cells indicated with yellow star. (C) An example of mitotic progression following DMSO control vehicle treatment. Red and yellow dashed lines indicate two daughter cells. Nocodazole or DMSO added at the beginning of imaging (0 hr 0 min). Scale bars, 10 μm.
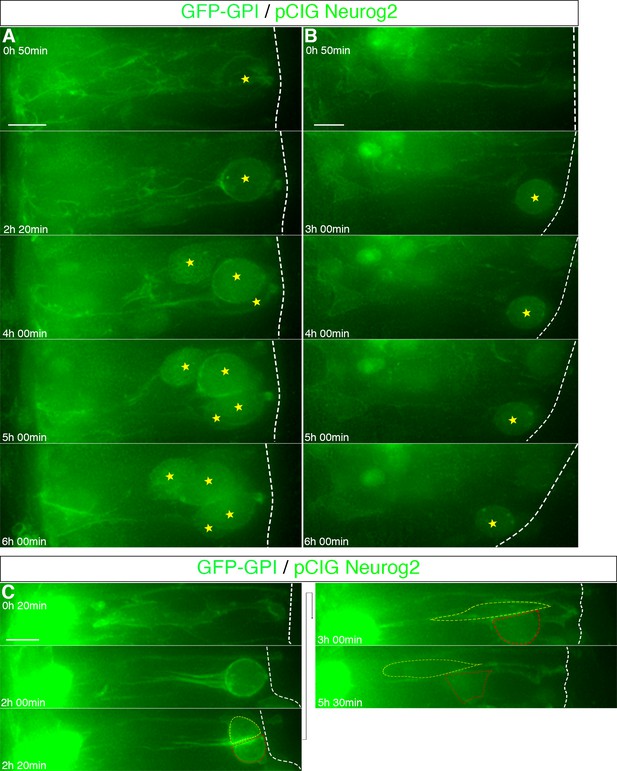
Assessment of taxol treatment efficacy in neural tube slices.
(A–B) Two examples of mitotic arrest following taxol treatment. Arrested cells indicated with yellow star. (C) An example of mitotic progression following DMSO vehicle control treatment. Red and yellow dashed lines indicate two daughter cells. Taxol or DMSO added at the beginning of imaging (0 hr 0 min). Scale bars, 10 μm.
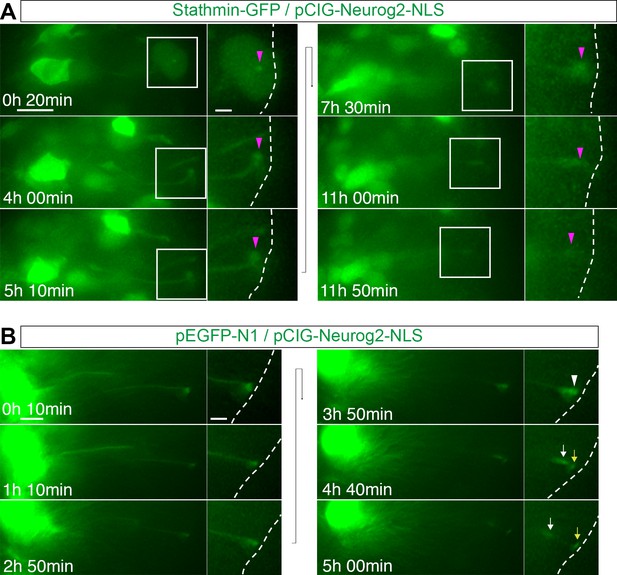
Stathmin-GFP mis-expression results in reduced delamination.
(A) TIme lapse sequence of a cell mis-expressing Stathmin-GFP (green) and pCIG-Neurog2 (nuclear, green). (B) Time lapse of a cell mis-expressing GFP and pCIG-Neurog2 (nuclear, green).
Time-lapse sequence of cells imaged in medium containing DMSO-vehicle control; this video is related to Figure 5A.
Apical abscission is promoted by the mis-expression of Neurog2 (pCIG-Neurog2) that labels the nucleus (green), GFP-GPI labels the cell membrane (green). Maximum intensity projections of deconvolved image sequences were used. Images acquired every 10 min. Boxed region magnified.
Time-lapse sequence of cells imaged in medium containing nocodazole; this video is related to Figure 5B.
Apical abscission is promoted by the mis-expression of Neurog2 (pCIG-Neurog2) that labels the nucleus (green), GFP-GPI labels the cell membrane (green). Maximum intensity projections of deconvolved images were used. Images acquired every 10 min.
Time-lapse sequence of cells imaged in medium containing nocodazole; this video relates to Figure 5—figure supplement 1B.
Cell membrane is labeled with GFP-GPI (green). Maximum intensity projections of deconvolved images were used. Images acquired every 10 min.
Time-lapse sequence of cells imaged in medium containing DMSO vehicle control; this video relates to Figure 5—figure supplement 1C.
Cell membrane is labeled with GFP-GPI (green). Maximum intensity projections of deconvolved images were used. Images acquired every 10 min.
Time-lapse sequence of cells imaged in medium containing taxol; this video relates to Figure 5—figure supplement 2A.
Cell membrane is labeled with GFP-GPI (green) and the nucleus with Neurog2 (pCIG-Neurog2). Maximum intensity projections of deconvolved images were used. Images acquired every 10 min.
Time-lapse sequence of cells imaged in medium containing taxol; this video is related to Figure 5C.
Apical abscission is promoted by the mis-expression of Neurog2 (pCIG-Neurog2) that labels the nucleus (green), GFP-GPI labels the cell membrane (green). Maximum intensity projections of deconvolved images were used. Images acquired every 10 min.
Time-lapse sequence of cell dynamics following Stathmin-GFP mis-expression; this video is related to Figure 5—figure supplement 3A.
Apical abscission is promoted by mis-expression of Neurog2 (pCIG-Neurog2) that labels the nucleus (green), Stathmin-GFP labels the cytoplasm (green). Maximum intensity projections of deconvolved images were used. Images acquired every 10 min.
Time-lapse sequence of cell dynamics following pEGFP-N1 mis-expression; this video is related to Figure 5—figure supplement 3B.
Apical abscission is promoted by mis-expression of Neurog2 (pCIG-Neurog2) that labels the nucleus (green), pEGFP-N1 labels the cytoplasm (green). Maximum intensity projections of deconvolved images were used. Images acquired every 10 min.
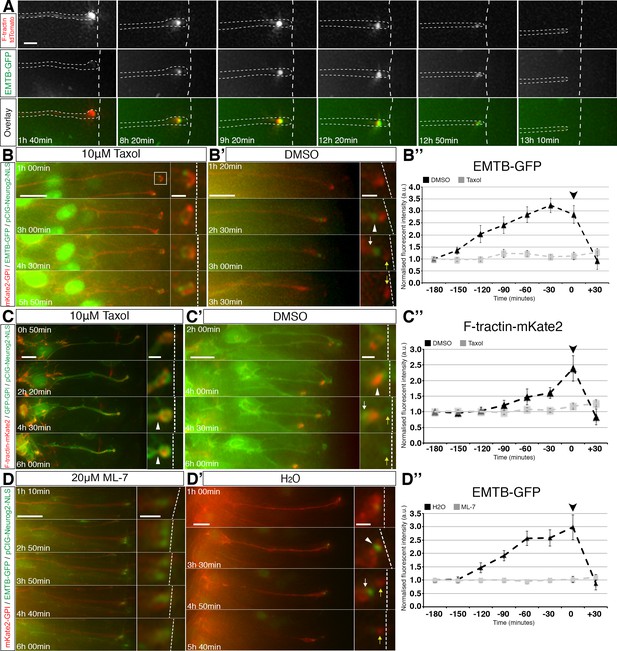
Coordination of sub-apical actin and microtubule dynamics.
(A) Live imaging of sub-apical actin (F-tractin td-Tomato) and microtubule (EMTB-GFP) dynamics during apical abscission. (B–B’, C–C’, D–D’). Time-lapse sequences of neural tube in embryo slices electroporated with EMTB-GFP/pCIG-Neurog2/mKate2 GPI or F-tractin-mKate2/pCIG-Neurog2/GFP GPI and treated with taxol (B, C) or ML-7 (D) or control vehicle (B’, C’, D’). Abscission site (white arrowheads), withdrawing apical process (white arrows), abscised particle (yellow arrows) and apical side (white dashed line) (B’’, C’’, D’’) Line graphs of normalised fluorescence intensities of EMTB-GFP or F-tractin-mKate2 dynamics in taxol or ML-7 (grey dashed line) and their control vehicles (black dashed line), quantified for 3 hr 30 min at 30 min intervals. EMTB-GFP dynamics are significantly affected by the taxol and ML-7 treatment (2-way ANOVA, p<0.001 for each of the treatments, error bars = SEM). F-tractin-mKate2 dynamics are significantly affected by ML-7 treatment (2-way ANOVA, p=0.002, error bars = SEM). Black arrowhead is abscission point for controls. Scale bars, (A) 2 μm, (B–B’) (C–C’) (D–D’) 10 μm; enlarged regions, 2 μm.Figure
-
Figure 6—source data 1
Quantification of EMTB-GFP and F-tractin-mKate2 fluorescence.
- https://doi.org/10.7554/eLife.26215.034
Time-lapse sequence of actin and microtubule dynamics during apical abscission; this video is related to Figure 6A.
Actin is labelled with F-tractin-td-Tomato (red) and microtubules with EMTB-GFP (green) during apical abscission. Apical abscission is promoted by mis-expression of Neurog2 (pCIG-Neurog2) that labels the nucleus (green). Maximum intensity projections of deconvolved images are used. Images acquired every 10 min. Only the apical region is shown in the video.
Time-lapse sequence of microtubule dynamics in cells imaged in medium containing taxol; this video is related to Figure 6B.
Mis-expressed Neurog2 (pCIG-Neurog2) labels the nucleus (green), microtubules are visualized with EMTB-GFP (green) and cell membrane with mKate2-GPI (red). Maximum intensity projections of deconvolved images are used. Images acquired every 10 min.
Time-lapse sequence of microtubule dynamics in cells imaged in medium containing DMSO vehicle control; this video is related to Figure 6B’.
Mis-expressed Neurog2 (pCIG-Neurog2) labels the nucleus (green), microtubules are visualized with EMTB-GFP (green) and cell membrane with mKate2-GPI (red). Maximum intensity projections of deconvolved images are used. Images acquired every 10 min.
Time-lapse sequence of actin dynamics in cells imaged in medium containing taxol; this video is related to Figure 6C.
Mis-expressed Neurog2 (pCIG-Neurog2) labels the nucleus (green), actin is visualized with F-tractin-mKate2 (red) and cell membrane with GFP-GPI (green). Maximum intensity projections of deconvolved images are used. Images acquired every 10 min.
Time-lapse sequence of actin dynamics in cells imaged in medium containing DMSO vehicle control; this video is related to Figure 6C’.
Mis-expressed Neurog2 (pCIG-Neurog2) labels the nucleus (green), actin is visualized with F-tractin-mKate2 (red) and cell membrane with GFP-GPI (green). Maximum intensity projections of deconvolved images are used. Images acquired every 10 min.
Time-lapse sequence of microtubule dynamics in cells imaged in medium containing ML-7; this video is related to Figure 6D.
Mis-expressed Neurog2 (pCIG-Neurog2) labels the nucleus (green), microtubules are visualized with EMTB-GFP (green) and cell membrane with mKate2-GPI (red). Maximum intensity projections of deconvolved images are used. Images acquired every 10 min.
Time-lapse sequence of microtubule dynamics in cells imaged in H2O vehicle control; this video is related to Figure 6D’.
Mis-expressed Neurog2 (pCIG-Neurog2) labels the nucleus (green), microtubules are visualized with EMTB-GFP (green) and membrane with mKate2-GPI (red). Maximum intensity projections of deconvolved images are used. Images acquired every 10 min.
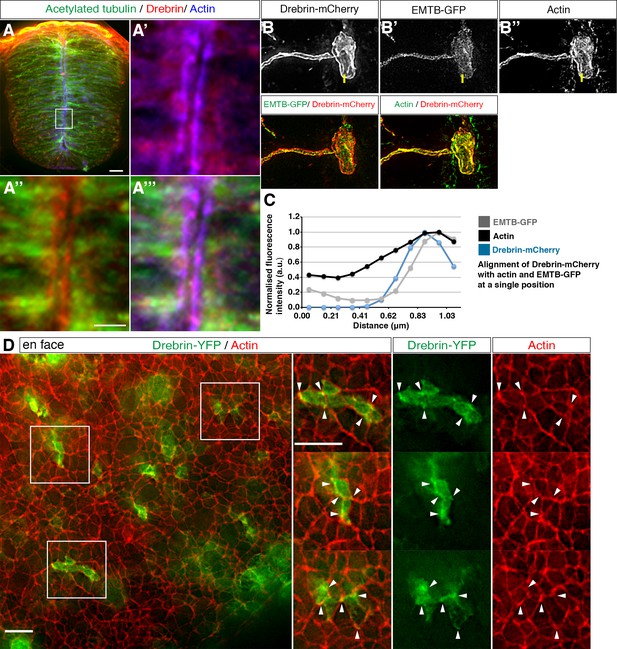
Drebrin localisation in the neural tube.
(A) Representative image of HH17-18 chick embryo neural tube labelled with antibodies to detect drebrin and acetylated α-tubulin and stained with phalloidin. Magnified boxed region shown in A'-A'''. End-foot of a neuroepithelial cell mis-expressing (B) Drebrin-mCherry and (B') EMTB-GFP and stained with (B'') phalloidin. (C) Representative line graphs of normalised fluorescence intensity at the level of the actin cable (B–B''). (D) En face imaging of neuroepithelial end-feet electroporated with Drebrin-YFP and stained for phalloidin. Boxed areas are magnified. White arrowheads indicate Drebrin-YFP and phalloidin co-localisation. Scale bars, (A) 20 μm and boxed region 5 μm, (B) 2 μm, (C) 10 μm.
-
Figure 7—source data 1
Drebrin-mCherry / Actin / EMTB-GFP alignment.
- https://doi.org/10.7554/eLife.26215.044
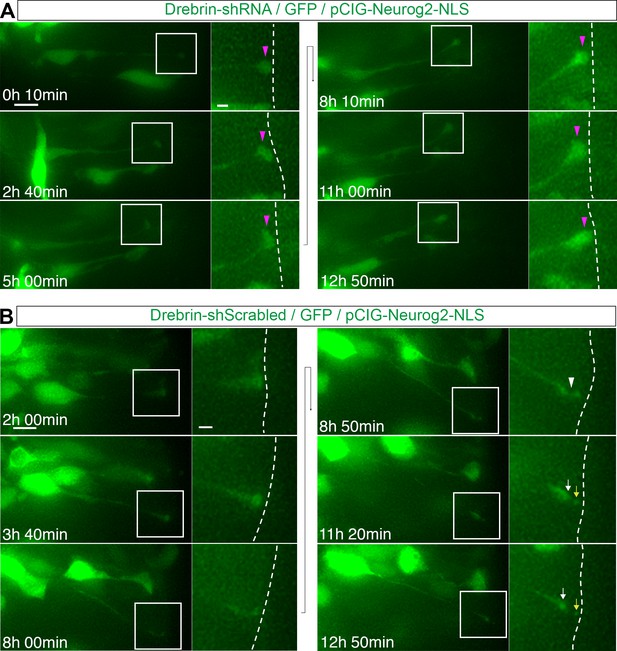
Drebrin knockdown reduces the incidence of delamination.
(A) Time lapse sequence of a cell mis-expressing Drebrin-shRNA (green) and pCIG-Neurog2 (nuclear, green). (B) Time lapse sequence of a cell mis-expressing Drebrin-shScrambled (green) and pCIG-Neurog2 (nuclear, green). Apical end process (purple arrowhead), abscission site (white arrowheads), withdrawing apical process (white arrows), abscised particle (yellow arrows) and apical side (white dashed line). Scale bars, 10 μm; enlarged regions, 2 μm.
Time-lapse sequence of cell behaviour following Drebrin knockdow; this video is related to Figure 7—figure supplement 1A.
Neuronal delamination is promoted by the mis-expression of Neurog2 (pCIG-Neurog2) that labels the nucleus (green), GFP expression from the Drebrin-shRNA construct labels the cytoplasm (green). Maximum intensity projections of deconvolved images were used. Images acquired every 10 min.
Time-lapse sequence of cell behaviour following scrambled shRNA construct expression; this video is related to Figure 7—figure supplement 1B.
Apical abscission is promoted by the mis-expression of Neurog2 (pCIG-Neurog2) that labels the nucleus (green), Drebrin-shScrambled labels the cytoplasm (green). Maximum intensity projections of deconvolved images were used. Images acquired every 10 min.
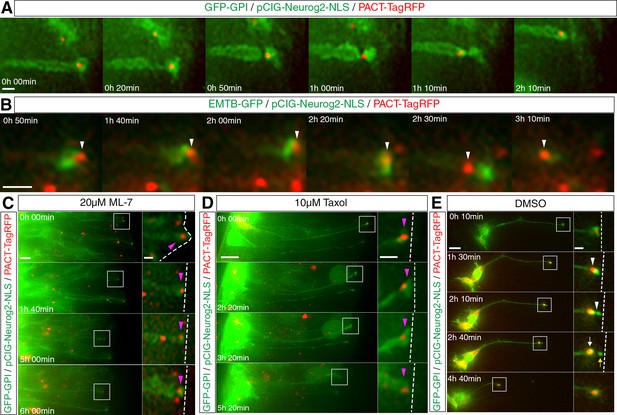
Centrosome translocation during apical abscission depends on actin and microtubule dynamics.
(A) The centrosome (labelled with PACT-TagRFP, red) undergoes a basal translocation through a thinned region of membrane (labelled with GFP-GPI, green). (B) The centrosome (labelled with PACT-TagRFP, red) translocates through a condensed microtubule tunnel-like configuration (labelled with EMTB-GFP, green). (C–E) Time-lapse sequences of centrosome dynamics in cells imaged in medium containing ML-7 (C), taxol (D) or DMSO control (E). Embryo neural tubes electroporated with GFP-GPI/pCIG-Neurog2/PACT TagRFP. Apical end process (purple arrowhead), abscission site (white arrowheads), withdrawing apical process (white arrows), abscised particle (yellow arrows) and apical side (white dashed line). Scale bars, (A) (B) 2 μm, (C) (D) (E) 10 μm; enlarged regions, 2 μm.
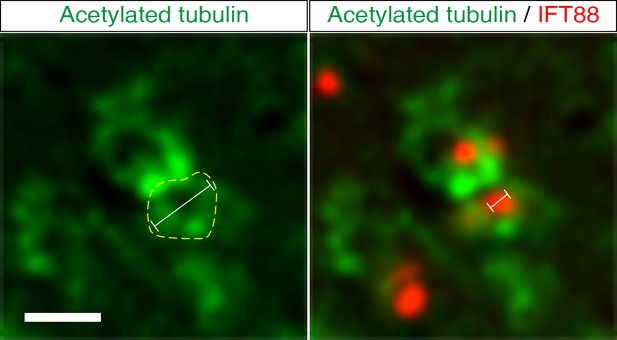
Measurement of apical microtubule rim and centrosome diameter.
The Z-stack section in which IFT88 appears largest was selected for measurement of acetylated tubulin ring (yellow dashed line around cell) and IFT88 labelled structure. Capped lines represent the diameter of the microtubule ring and IFT88. Scale bar: 1 μm.
Time-lapse sequence of centrosome undergoing basal translocation; this video is related to Figure 8A.
Mis-expressed Neurog2 (pCIG-Neurog2) localises to the nucleus (green), the centrosome is visualized with PACT-TagRFP (red) and the cell membrane with GFP-GPI (green). Maximum intensity projections of deconvolved images are used. Images acquired every 10 min. Only the apical region is shown in the video.
Time-lapse sequence of centrosome translocation through a condensed microtubule tunnel-like configuration; this video is related to Figure 8B.
Mis-expressed Neurog2 (pCIG-Neurog2) localises to the nucleus (green), the centrosome is visualised with PACT-TagRFP (red) and microtubules with EMTB-GFP (green). Maximum intensity projections of deconvolved images are used. Images acquired every 10 min. Only the apical region is shown in the video. Arrowhead indicates the centrosome of interest.
Time-lapse sequence of centrosome dynamics in cells imaged in medium containing ML-7; this video is related to Figure 8C.
Mis-expressed Neurog2 (pCIG-Neurog2) labels the nucleus (green), the centrosome is visualised with PACT-TagRFP (red) and the cell membrane with GFP-GPI (green). Maximum intensity projections of deconvolved images are used. Images acquired every 10 min.
Time-lapse sequence of centrosome dynamics in cells imaged in medium containing taxol; this video is related to Figure 8D.
Mis-expressed Neurog2 (pCIG-Neurog2) localises to the nucleus (green), the centrosome is visualised with PACT-TagRFP (red) and the cell membrane with GFP-GPI (green). Maximum intensity projections of deconvolved images are used. Images acquired every 10 min.
Time-lapse sequence of centrosome dynamics in cells imaged in medium containing DMSO-vehicle control; this video is related to Figure 8E.
Mis-expressed Neurog2 (pCIG-Neurog2) localises to the nucleus (green), the centrosome is visualised with PACT-TagRFP (red) and the membrane with GFP-GPI (green). Maximum intensity projections of deconvolved images are used. Images acquired every 10 min.
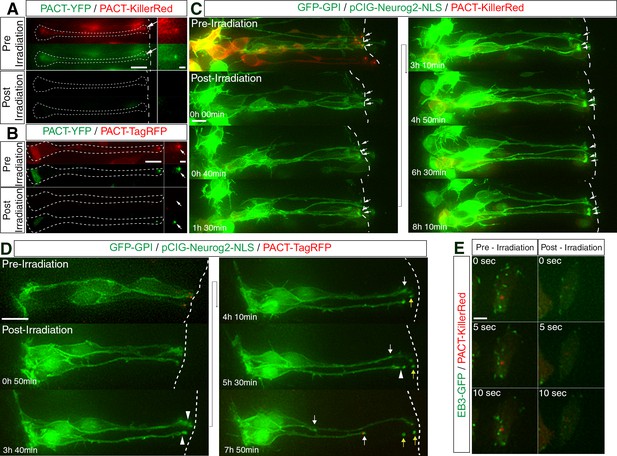
Compromised microtubule nucleating potential of the centrosome blocks delamination from the apical surface.
(A) Green light irradiation-mediated photobleaching of PACT-KillerRed, which localises to the centrosome, is accompanied by a corresponding depletion of PACT-YFP fluorescence. Arrows point to the centrosome, (B) Photobleaching of PACT-TagRFP following green light irradiation does not result in a corresponding reduction of PACT-YFP fluorescence. Arrows point to the centrosome. (C) Time-lapse sequence of neural progenitors following CALI. Cells poised to differentiate remain attached to the apical surface. Cells were electroporated with pCIG-Neurog2, GFP-GPI and PACT-KillerRed. White arrows point to the apical tips of cells that have been subjected to CALI. (D) Time-lapse sequence of neural progenitors following the imaging regime used for CALI. Two out of three cells underwent apical abscission during the 8 hr post-irradiation imaging period. Cells were electroporated with pCIG-Neurog2, GFP-GPI and PACT-TagRFP in place of the PACT-KillerRed construct. Abscission site (white arrowheads), withdrawing apical process (white arrows), abscised particle (yellow arrows) and apical side (white dashed line). (E) Reduction in the microtubule nucleation potential (48% reduction) of the centrosome, 3 hr post-CALI. Stills of EB3-GFP comets pre- and post-irradiation of a single end-foot (en face). Scale bars, (A–D) 10 μm; enlarged regions, 2 μm, (E) 2 μm.
Time-lapse sequence of cells following green-light irradiation; this video is related to Figure 9B.
The centrosome is labelled with both PACT-TagRFP (red) and PACT-YFP (green). First timepoint is when PACT-TagRFP is visible before beginning the imaging regime for CALI. Maximum intensity projections of deconvolved images are used. Images acquired every 5 min.
Time-lapse sequence of cell behaviour following CALI; this video is related to Figure 9C.
PACT-KillerRed localises to the PCM. Apical abscission is promoted by mis-expression of Neurog2 (pCIG-Neurog2) that labels the nucleus (green) and cell membrane is labelled with GFP-GPI (green). Maximum intensity projections of deconvolved images are used. Images acquired every 10 min.
Time-lapse sequence of cell behaviour following the CALI green light irradiation; this video is related to Figure 9D.
PACT-TagRFP localises to the PCM. Apical abscission is promoted by mis-expression of Neurog2 (pCIG-Neurog2) that labels the nucleus (green) and cell membrane is labelled with GFP-GPI (green). Maximum intensity projections of deconvolved images are used. Images acquired every 10 min.
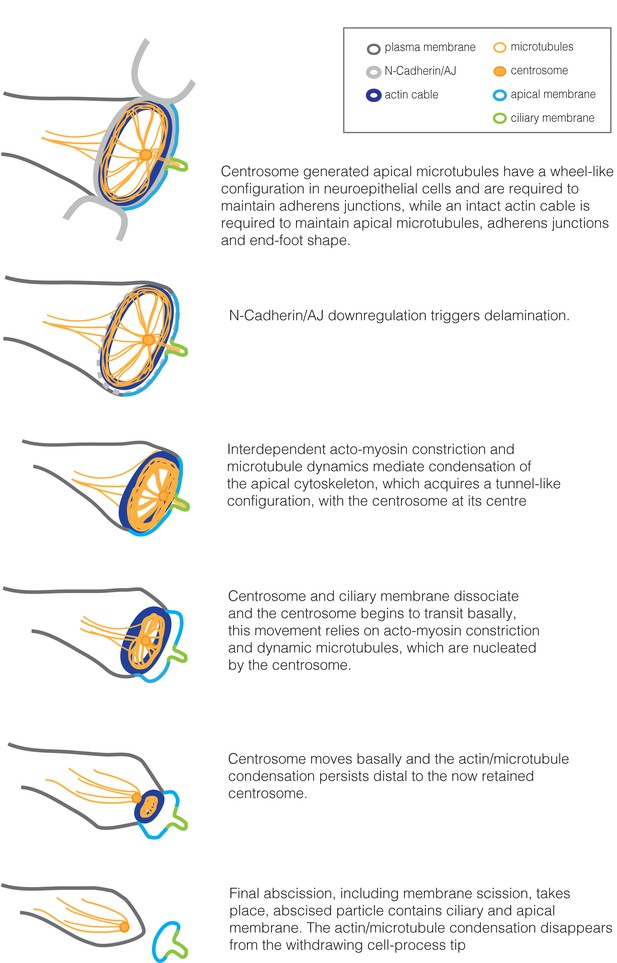
Summary of cytoskeletal configuration and dynamic changes in the apical end-foot during neuronal delamination.
https://doi.org/10.7554/eLife.26215.058Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.26215.059





