Homeostatic control of START through negative feedback between Cln3-Cdk1 and Rim15/Greatwall kinase in budding yeast
Figures
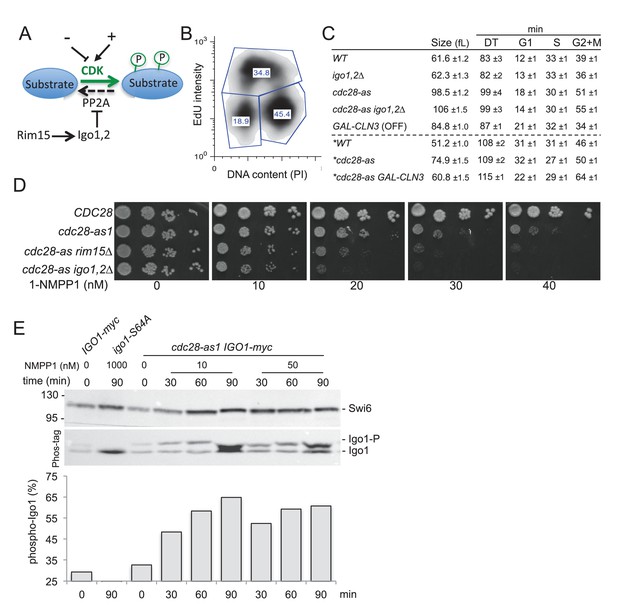
The Rim15 pathway is activated and becomes essential when Cdc28 activity is low.
(A) The Rim15 pathway can potentiate CDK-dependent phosphorylation of target proteins by inhibiting the CDK-counteracting PP2A phosphatase. (B) Bivariate EdU/PI FACS profile of asynchronous WT cells (E3087) pulsed for 10 min with 25 µM EdU. The proportion of G1 (lower left polygon), S phase (upper polygon) and G2+M cells (lower right polygon) is indicated. (C) Median cell volume (in fL), doubling time (DT in min) and the duration of the G1, S and G2+M phases (in min) is indicated for each strain grown in SCD or SCRaf+Gal (asterisk) medium at 30°C in the absence of 1-NMPP1 (E3087, E4259, E4479, E4458, E5493, E5492). Measures were done in triplicate and represented as Mean ± SEM. (D) Cells of the indicated genotypes (E3087, E4479, E4471, E4458) were spotted in fivefold serial dilution on YPD plates containing increasing concentrations of 1-NMPP1. Rim15 and Igo1,2 become essential for viability in cdc28-as1 cells exposed to more than 20 nM 1-NMPP1. (E) Analysis of Rim15 activity by monitoring the level of Igo1 phosphorylation on Phos-tag gels. IGO1-myc, igo1-S64A-myc and cdc28-as1 IGO1-myc cells (E4974, E4975 and E4996, respectively) were exposed for 0, 30, 60 or 90 min to 10 or 50 nM 1-NMPP1 and Igo1 phosphorylation was examined by western blotting of Phos-tag gels. Quantification of the fraction of Igo1-S64 phosphorylation is shown below. Swi6, loading control.
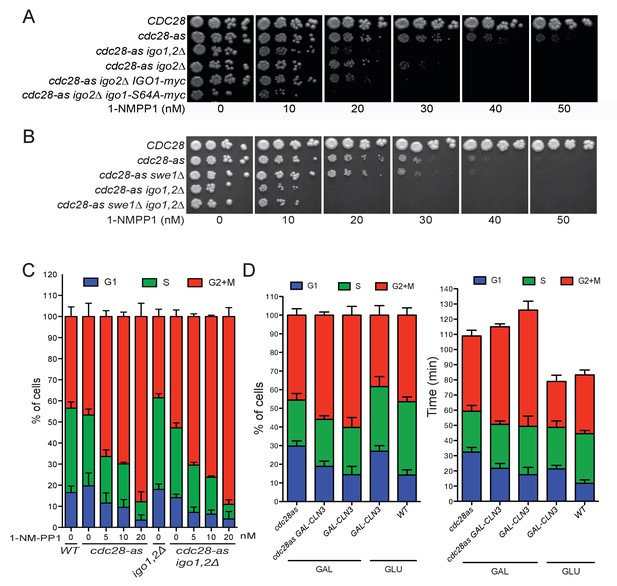
Hypersensitivity of cdc28-as1 igo1,2∆ cells is due to G1, not G2, defect.
(A) Non-phosphorylable igo1-S64A phenocopies an igo1∆ deletion for hypersensitivity to 1-NMPP1. Cells of the indicated genotype (E3087, E4479, E4458, E4331, E4496, E4495) were spotted in fivefold serial dilution on YPD plates containing increasing concentrations of 1-NMPP1. (B) Hypersensitivity of cdc28-as1 igo1,2∆ cells to 1-NMPP1 is independent of SWE1. Strains E3087, E4479, E4452, E4458, E5169. (C) Cell cycle distribution determined by EdU-PI bivariate FACS analysis of WT (E3087), cdc28-as1 (E4479) igo1,2∆ (E4259) and cdc28-as1 igo1,2∆ (E4458) cells grown in SC-D containing the indicated concentrations of 1-NMPP1. (D) Cell cycle distribution (left) and cycle phase duration (right) of WT (E3087), cdc28-as1 (E4479), GAL-CLN3 (E5493) and cdc28-as1 GAL-CLN3 (E5492) cells grown at 30°C in SC-D or SC-Gal.
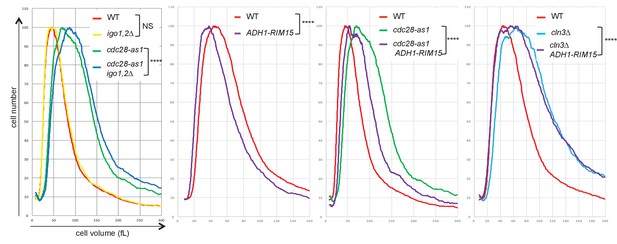
The Rim15-Igo1,2 pathway controls cell size.
Cell size distribution of asynchronous populations of cells growing in SC-D medium at 30°C was analysed on a CASY 1-TTC cell analyser (>10,000 cells analysed/sample). Pairwise comparisons indicate that wild-type and igo1,2∆ populations are not significantly different (NS), whereas all other comparisons show differences in cell size distributions (Chi2 test, p<0.0001; ****), with igo1,2∆ ablation causing size increase in cdc28-as1 cells (left panel) and RIM15 overexpression causing cell size reduction in WT, cdc28-as1 and cln3∆ cells. Strains: wild-type (E3087), igo1,2∆ (E4255), cdc28-as1 (E4479), cdc28-as1 igo1,2∆ (E4458), ADH1-RIM15 (E5504), cdc28-as1 ADH1-RIM15 (E5323), cln3∆ (E5493) and cln3∆ ADH1-RIM5 (E5505).
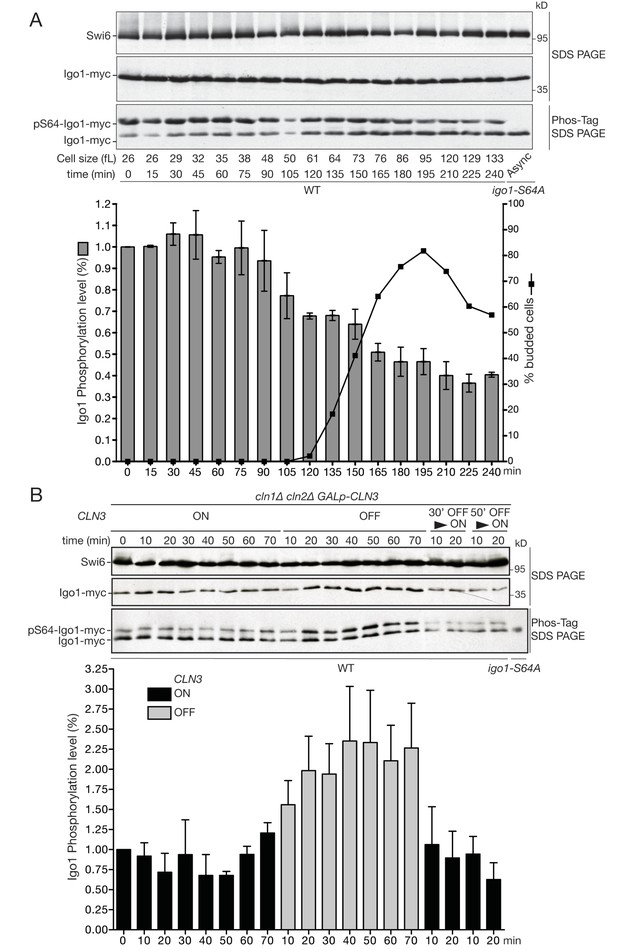
The Rim15 pathway is active in early G1 cells and negatively regulated by Cln3-Cdk1 kinase.
(A) Western blot and quantification of the level of Igo1 phosphorylation (P-Igo1/total Igo1) during a synchronous cell cycle after elutriation of IGO1-myc cells (E4996). Mean (SD) of three experiments. Value was arbitrarily set at 1 for the first time point. (B) Igo1-S64 phosphorylation level in exponentially growing cln1∆ cln2∆ cln3::GAL-CLN3 cells (E5261) shifted from SC-Gal to SC-D for the indicated time, filtered after 30 or 50 min and put back in SC-Gal for 10 and 20 min. Quantification of phospho-S64-Igo1 shows the mean (SD) of three experiments. Value was arbitrarily set at 1 for the first time point.
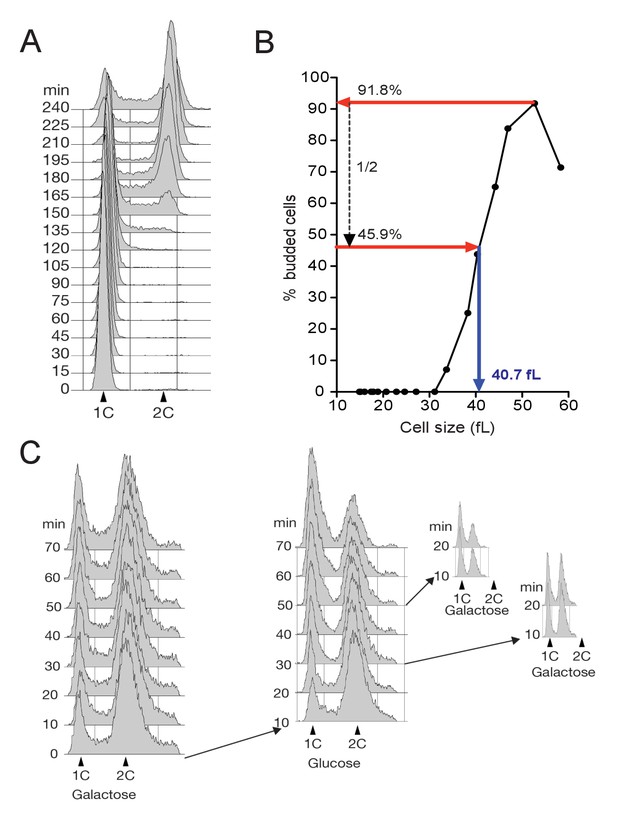
FACS profiles and determination of median cell size at budding
(A) Cell cycle progression determined by FACS analysis of DNA content of elutriated early IGO1-myc cells (E4996) released in SC-D at 30°C (corresponding to Figure 2A). (B) Calculation of cell size at half-maximal budding. Median cell volume and budding index are measured at each time point during a synchronous cell cycle after elutriation. Normalized cell size at budding is calculated as the median cell size when the population reaches half-maximum budding. (C) FACS profiles of experiment shown in Figure 2B.
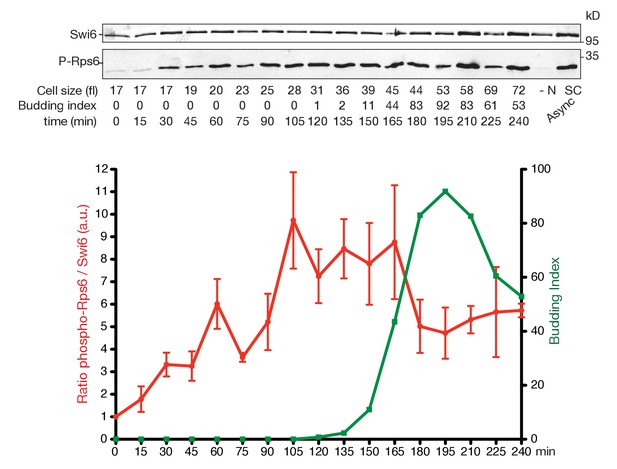
TORC1 activity increases during G1 and drops in mitosis.
Rps6 phosphorylation, a proxy for TORC1 activity, was quantitated throughout the cell cycle of WT cells (E3087) synchronized by centrifugal elutriation. Top panel: western blot of Swi6 (loading control) and phospho-Rps6 (P-Rps6). The two rightmost lanes are asynchronous cells grown in SC-D medium (SC) or starved for nitrogen (-N) for 1 hr (YNB w/o NH4SO4, 2% D). Bottom panel: quantification of P-Rps6 relative to Swi6 (red) showing mean and SD of three technical duplicates. Budding index (green).
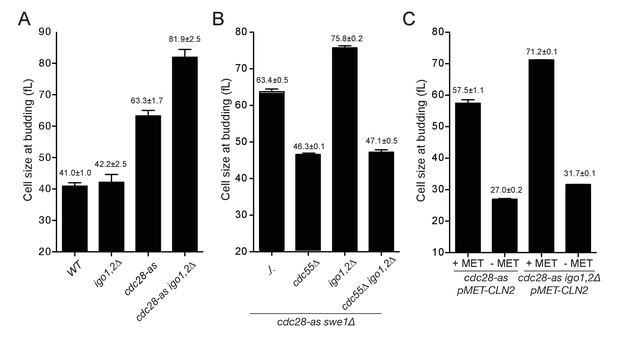
Rim15 and Cln-Cdk1 kinases cooperate to set the cell size at START.
(A) Cells lacking Igo1,2 bud at a larger cell size. Elutriated early G1 cells of the indicated genotypes (E3087, E4259, E4479, E4458) were inoculated in SC-D medium at 30°C, and the population modal cell volume measured at the time of half-maximum budding. (B) The START delay of cdc28-as1 igo1,2∆ cells depends on CDC55. All strains (E4465, E5169, E4452, E4463) are also deleted for SWE1. (C) Ectopic CLN2 expression from the MET3 promoter suppresses the START delay of cdc28-as1 and cdc28-as1 igo1∆ igo2∆ cells. Early G1 cdc28-as1 pMET3-CLN2 (E5447) and cdc28-as1 igo1∆ igo2∆ pMET3-CLN2 (E5159) cells were obtained by elutriation and put back in SC-D medium containing methionine (CLN2 off) or not (CLN2 on), and their size at budding measured as above. Cell volumes are indicated (fL) and are the mean (±SEM) of three independent elutriations for each strain.
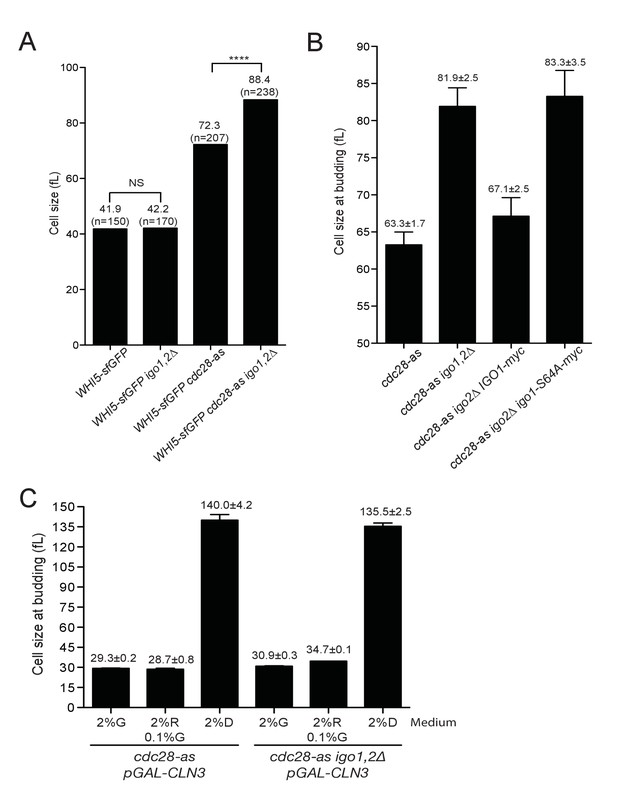
Rim15 and Cln-Cdk1 kinases cooperate to set the cell size at START.
(A) Single cell measurements of cell size in G1. Median cell volume of Whi5-positive unbudded cells in asynchronously growing cells of the indicated genotype (E5100, E4948, E5101, E5102), determined with BudJ software. N indicates the number of cells measured in each strain. Mann-Whitney test was used to determine if medians are different (NS, non-significant). (B) Increased cell size at budding in cdc28-as1 igo1,2∆ cells is due to Rim15. Median cell size at budding after elutriation of the indicated strains (E4479, E4458, E4996, E4995) released in SC-D at 30°C. Mean ±SD of two independent experiments. (C) Cell size at budding of cdc28-as1 and cdc28-as1 igo1,2∆ depends on CLN3. Size at budding after elutriation was measured for cdc28-as1 (E4479), cdc28-as1 igo1,2∆ (E4458), cdc28-as1 GAL-CLN3 (E5373) and cdc28-as1 igo1,2∆ GAL-CLN3 (E5231) grown SC-D, SC-Gal or SC-Raf +0.1% Gal. Mean ±SD of two independent experiments.
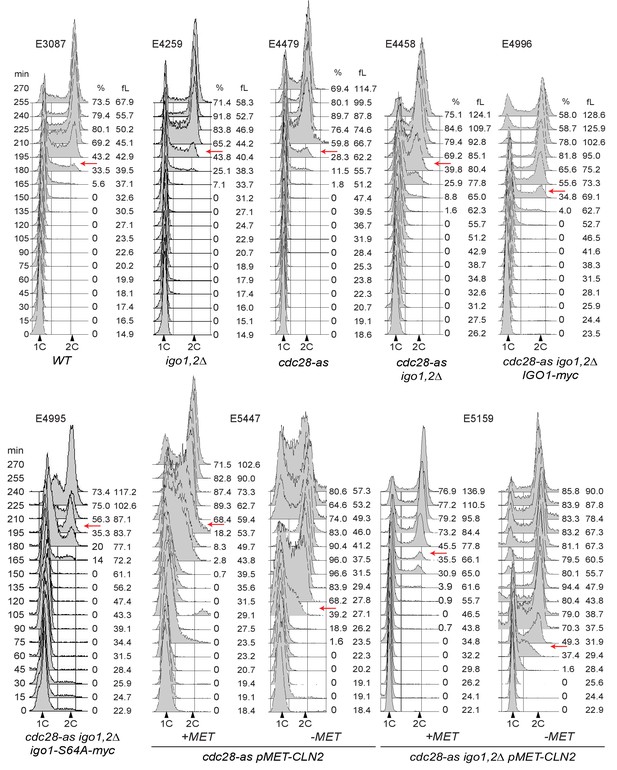
Cell cycle progression determined by FACS analysis of DNA content of elutriated early cells of the indicated genotype released in SC-D at 30°C.
The peak cell size (in fL) and the budding index are given for each time point. BI, Budding index. The red arrow indicates when half of cells have budded. Elutriations were done three to six times independently for each strain; one example is shown (corresponding to Figure 3A–C). Strain number is indicated atop each histogram.
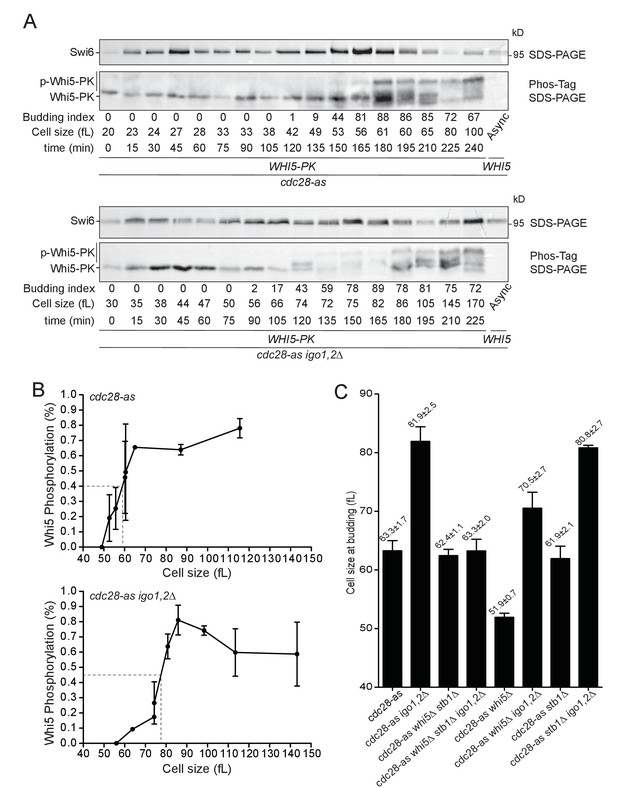
The Rim15 pathway controls START by promoting Whi5 phosphorylation.
(A) Whi5 is phosphorylated at a larger cell size when Igo1,2 are missing. cdc28-as1 (E5443) and cdc28-as1 igo1,2∆ (E5441) containing PK-tagged Whi5 were elutriated and Whi5 phosphorylation timing determined on Phos-tag gels. Untagged WHI5 strain is shown as negative control for antibody specificity. (B) Quantification of Whi5 phosphorylation from Phos-tag western blots. Mean (SD), n = 3. (C) The START delay of cdc28-as1 igo1,2∆ cells is dependent on WHI5 and STB1. Cells of the indicated genotype (E4479, E4458, E5222, E5218, E5188, E5227, E5189, E5221) were elutriated, early G1 cells incubated in SC-D at 30°C and the mean cell size at half maximum budding measured. Mean ±SD is indicated, n = 4.
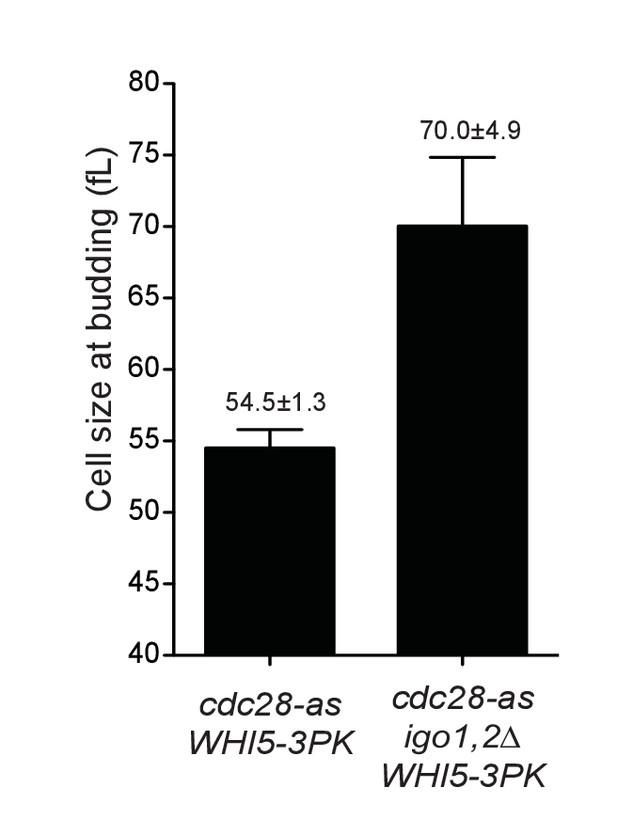
Cell size at budding after elutriation of cdc28-as1 WHI5-3PK (E5443) and cdc28-as1 igo1,2∆ WHI5-3PK (E5441) cells grown in SC-D.
Mean ±SD of four experiments (corresponding to Figure 4A).
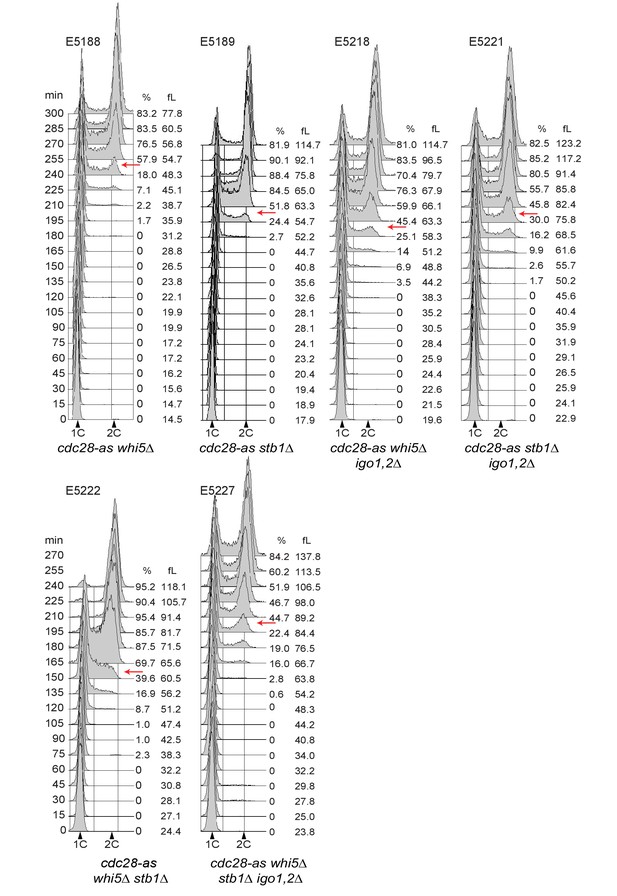
Cell cycle progression determined by FACS analysis of DNA content of elutriated early cells of the indicated genotype released in SC-D at 30°C.
The peak cell size (in fL) and the budding index are given for each time point. BI, Budding index. The red arrow indicates when half of cells have budded. Elutriations were done three to six times independently for each strain; one example is shown (corresponding to Figure 4C).
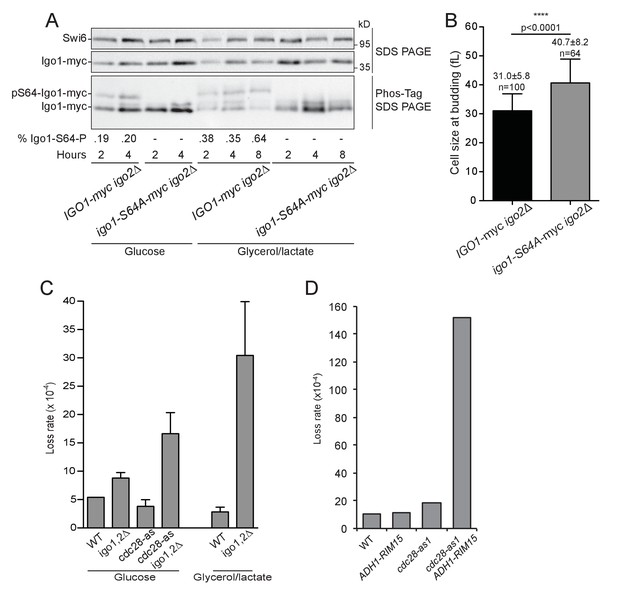
The Rim15 pathway is activated after the diauxic shift where it contributes to genome stability.
(A) The level of Igo1 phosphorylation in IGO1-myc (E4974) and igo1S64A-myc (E4975) cells growing either on glucose or glycerol-lactate was determined on Phos-tag gels. Cells were shifted for 2 to 8 hr to YEPD (Glucose) or YEPGL (Glycerol/lactate) and whole cell protein extract (10 µg) analysed by SDS-PAGE or Phos-tag-PAGE. Cells grew longer in YEPGL (8 hr) so to reach the same cell density as in 4 hr in YEPD. The fraction of phospho-S64-Igo1, shown below the gel, was quantitated as in Figure 2. Swi6, loading control. (B) Cells containing non-phosphorylatable Igo1 pass START at a larger size when grown in respiratory conditions. IGO1 and igo1-S64A cells grown in YEP-Glycerol/lactate were fixed, imaged and their size at budding determined using BudJ software. Mean cell volume ± SEM, n = 100 and n = 64, respectively; Student t test. (C) Chromosome loss in igo1,2∆ mutants. Cells of the indicated genotype (E4989, E4990, E5003, E5001) containing a 61 kb circular chromosome harbouring three ARSs (RCIII-3ARS) were grown for 6–8 generations without selection and then plated to determine the fraction of cells having lost RCIII (red colonies). Cells were grown either in SC-D (left) or in YEP-Lactate/Glycerol (right). Numbers indicate loss rate /cell • generation (Mean ± SEM, n = 3, 5–8000 colonies/sample). (D) Chromosome loss in ADH1-RIM15 cells. Cells of shown genotype (E4989, E5497, E5003, E5498) were grown in SC-D for 12–15 generations and plated on sectoring medium to score red colonies having lost the RCIII chromosome, as in (C).
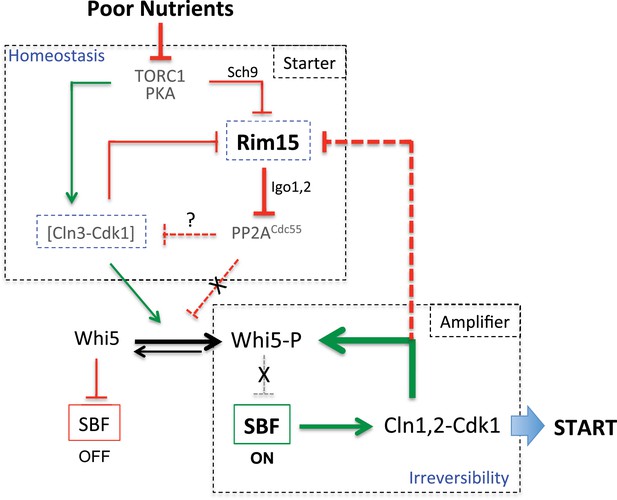
Model for homeostatic control of START by antagonism between Cln3-Cdk1 and Rim15, and cooperation for Whi5 phosphorylation.
TORC1, PKA and PP2ACdc55 have additional targets while Cln3-Cdk1 also targets Stb1, which are not shown here for simplicity. When nutrients are poor, TORC1 is down regulated leading to low Cln3 levels and higher Rim15 activity. High Rim15 then inhibits PP2ACdc55 to promote Whi5 phosphorylation and START despite low Cln3-Cdk1 activity. Conversely, when nutrients are rich, high TORC1 leads to Cln3 accumulation, Rim15 inhibition and high PP2ACdc55 activity. The regulatory network leading to START and cell cycle entry can be decomposed in two modules: the Starter module (on top) favours initial Whi5 phosphorylation by decreasing PP2A activity when Cln3-Cdk1 activity is low, is coupled to nutrient availability and provides cell size homeostasis; the Amplifier module (on bottom) brings in positive feedback by Cln1,2-Cdk1 for full Whi5 phosphorylation and irreversibility of the START transition. Cln3-Cdk1 is inactivated by dephosphorylation (Tyers et al., 1992), possibly by PP2A. Negative feedback between Cln3-Cdk1 and Rim15 in the homeostatic module maintains a proper balance between kinase and phosphatase activities acting on Whi5.
Tables
Yeast strains.
| Name | Genotype | |||||
|---|---|---|---|---|---|---|
| E3087 | MATa, URA3::GPD-TK(5x), AUR1c::ADH-hENT1 | |||||
| E4259 | MATa, igo1::NatNT, igo2::KanMX, URA3::GPD-TK(5x), AUR1c::ADH-hENT1 | |||||
| E4331 | MATa, cdc28-as1(F88G), igo2::KanMX, URA3::GPD-TK(5x), AUR1c::ADH-hENT1 | |||||
| E4452 | MATa, cdc28-as1(F88G), swe1::LEU2, URA3::GPD-TK(5x), AUR1c::ADH-hENT1 | |||||
| E4458 | MATa, cdc28-as1(F88G), igo1::NatNT, igo2::KanMX, URA3::GPD-TK(5x), AUR1c::ADH-hENT1 | |||||
| E4463 | MATa, cdc28-as1(F88G), swe1::LEU2, igo1::NatNT, igo2::KanMX, URA3::GPD-TK(5x), AUR1c::ADH-hENT1 | |||||
| E4465 | MATa, cdc28-as1(F88G), swe1::LEU2, cdc55::TRP1, URA3::GPD-TK(5x), AUR1c::ADH-hENT1 | |||||
| E4471 | MATa, cdc28-as1(F88G), rim15::KanMX, URA3::GPD-TK(5x), AUR1c::ADH-hENT1 | |||||
| E4479 | MATa, cdc28-as1(F88G), URA3::GPD-TK(5x), AUR1c::ADH-hENT1 | |||||
| E4948 | MATα, igo1::NatNT, igo2::KanMX, WHI5-sfGFP | |||||
| E4974 | MATa, igo1::NatNT, igo2::KanMX, TRP1::IGO1-myc8, URA3::GPD-TK(5x), AUR1c::ADH-hENT1 | |||||
| E4975 | MATa, igo1::NatNT, igo2::KanMX, TRP1::IGO1-S64A-myc8, URA3::GPD-TK(5x), AUR1c::ADH-hENT1 | |||||
| E4989 | MATα, RCIII-SUP11-LEU2-3ARS | |||||
| E4990 | MATα, igo1::NatNT, igo2::KanMX, RCIII-SUP11-LEU2-3ARS | |||||
| E4995 | MATa, cdc28-as1(F88G), igo1::NatNT, igo2::KanMX, TRP1::igo1-S64A-myc8, URA3::GPD-TK(5x), AUR1c::ADH-hENT1 | |||||
| E4996 | MATa, cdc28-as1(F88G), igo1::NatNT, igo2::KanMX, TRP1::IGO1-myc8, URA3::GPD-TK(5x), AUR1c::ADH-hENT1 | |||||
| E5001 | MATα, cdc28-as1(F88G), igo1::NatNT, igo2::KanMX, RCIII-SUP11-LEU2-3ARS | |||||
| E5003 | MATα, cdc28-as1(F88G), RCIII-SUP11-LEU2-3ARS | |||||
| E5100 | MATα, WHI5-sfGFP | |||||
| E5101 | MATα, cdc28-as1(F88G), WHI5-sfGFP, URA3::GPD-TK(5x), AUR1c::ADH-hENT1 | |||||
| E5102 | MATα, cdc28-as1(F88G), igo1::NatNT, igo2::KanMX, WHI5-sfGFP, URA3::GPD-TK(5x), AUR1c::ADH-hENT1 | |||||
| E5159 | MATa, cdc28-as1(F88G), igo1::NatNT, igo2::KanMX, TRP1::MET3-CLN2(1x) | |||||
| E5169 | MATa, cdc28-as1(F88G), swe1::LEU2, cdc55::TRP1, igo1::NatNT, igo2::KanMX, | |||||
| E5188 | MATα, cdc28-as1(F88G), whi5::KanMX, AUR1c::ADH-hENT1, URA3::mCherry-TUB1 | |||||
| E5189 | MATα, cdc28-as1(F88G), stb1::KanMX, URA3::GPD-TK(5x) | |||||
| E5218 | MATa, cdc28-as1(F88G), whi5::KanMX, stb1::KanMX, igo1::NatNT, igo2::KanMX, URA3::GPD-TK(5x) | |||||
| E5221 | MATα, cdc28-as1(F88G), stb1::KanMX, igo1::NatNT, igo2::KanMX, URA3::GPD-TK(5x) | |||||
| E5222 | MATα, cdc28-as1(F88G), whi5::KanMX, stb1::KanMX, URA3::GPD-TK(5x) | |||||
| E5227 | MATa, cdc28-as1(F88G), whi5::KanMX, igo1::NatNT, igo2::KanMX, URA3::GPD-TK(5x) | |||||
| E5261 | MATa, igo1::NatNT, igo2::KanMX, TRP1::IGO1-myc8, cln1::hisG, cln2∆, cln3::GAL10-CLN3::URA3 | |||||
| E5231 | MATa, cdc28-as1(F88G), igo1::NatNT, igo2::KanMX, cln3::GAL10-CLN3::URA3 | |||||
| E5323 | MATα, cdc28-as1(F88G), rim15::NatNT::ADH-yeGFP-RIM15 | |||||
| E5373 | MATa, cdc28-as1(F88G), cln3::GAL10-CLN3::URA3 | |||||
| E5441 | MATa, cdc28-as1(F88G), igo1::NatNT, igo2::KanMX, WHI5-3PK::HIS3Kl, URA3::GPD-TK(5x), AUR1c::ADH-hENT1 | |||||
| E5443 | MATa, cdc28-as1(F88G), WHI5-3PK::HIS3Kl, URA3::GPD-TK(5x), AUR1c::ADH-hENT1 | |||||
| E5447 | MATa, cdc28-as1(F88G), TRP1::MET3-CLN2(1x) | |||||
| E5492 | MATa, cdc28-as1(F88G), cln3::GAL10-CLN3::URA3, URA3::GPD-TK(5x), AUR1c::ADH-hENT1 | |||||
| E5493 | MATa, cln3::GAL10-CLN3::URA3, URA3::GPD-TK(5x), AUR1c::ADH-hENT1 | |||||
| E5497 | MATa, rim15::NatNT::ADH-yeGFP-RIM15, RCIII-SUP11-LEU2-3ARS | |||||
| E5498 | MATa, cdc28-as1(F88G), rim15::NatNT::ADH-yeGFP-RIM15, RCIII-SUP11-LEU2-3ARS | |||||
| E5504 | MATa, rim15::NatNT::ADH-yeGFP-RIM15, URA3::GPD-TK(5x), AUR1c::ADH-hENT1 | |||||
| E5505 | MATa, rim15::NatNT::ADH-yeGFP-RIM15, URA3::GPD-TK(5x), AUR1c::ADH-hENT1, cln3::GAL10-CLN3::URA3 | |||||





