KLHL41 stabilizes skeletal muscle sarcomeres by nonproteolytic ubiquitination
Figures
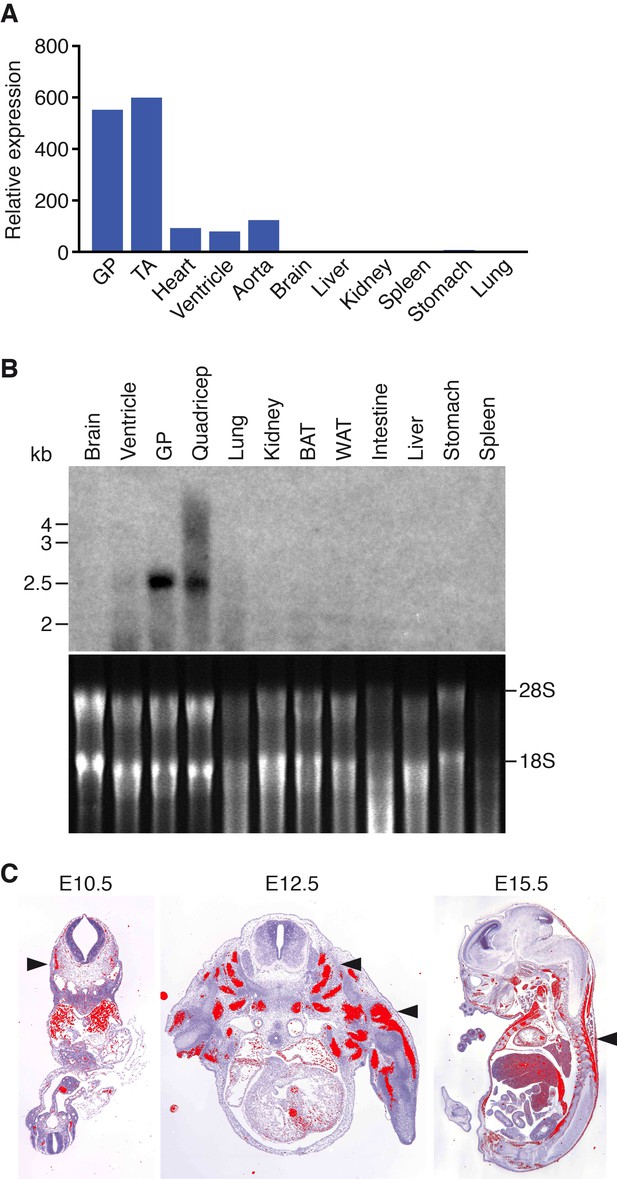
Muscle-specific expression of Klhl41.
(A) Distribution of Klhl41 transcript in adult mouse tissues as determined by qRT-PCR. Expression of Klhl41 transcript was normalized to 18 s rRNA. GP: Gastrocnemius plantaris, TA: Tibialis anterior. (B) Northern blot analysis of Klhl41 transcript in adult mouse tissues (top). Total RNA in each lane was visualized by ethidium bromide staining and 28 s and 18 s rRNAs were indicated on the right (bottom). BAT: Brown adipose tissue, WAT: White adipose tissue. (C) Detection of Klhl41 transcript by in-situ hybridization using an Klhl41 anti-sense radioisotopic probe in mouse embryo sections at indicated ages. Transverse sections of E10.5 and E12.5 embryos, and sagittal section of an E15.5 embryo are shown. Signal for Klhl41 mRNA (pseudocolored red) only appears in developing muscle. Arrowheads point to representative developing skeletal muscle. The low signal outside the developing muscles represents background.
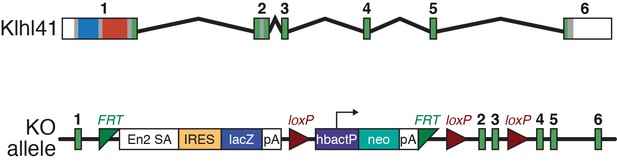
Klhl41 KO allele.
Structure of Klhl41 WT (top) and KO allele (bottom). Insertion of the LacZ cassette into intron 1 is sufficient to abolish Klhl41 expression.
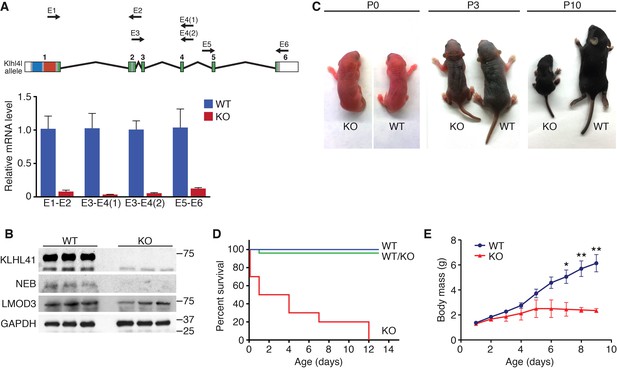
Klhl41 KO mice display neonatal lethality.
(A) Klhl41 gene structure: the regions coding for BTB (blue), BACK (red) and Kelch repeats (green) are indicated (top). Detection of Klhl41 transcript in P0 muscle of WT and Klhl41 KO mice by qRT-PCR using the indicated primer pairs (arrows) (bottom) (n = 3 mice per genotype). Data are presented as mean ±SEM. (B) Western blot analysis of the indicated proteins in WT and KO hindlimb muscles from P0 pups. Because of the large size of NEB (800 > KDa), the corresponding band is above the molecular weight markers used. GAPDH was used as a loading control. (C) Surviving WT and Klhl41 KO mice at P0, P3 and P10. KO pups show a failure-to-thrive phenotype and become severely runted by 10 days of age. (D) Survival curve of offspring from Klhl41 heterozygous intercrosses. n = 20 WT, n = 10 WT/KO (heterozygous), and n = 10 KO. (E) Growth curves (body mass) of WT and Klhl41 KO mice. n = 20 WT and n = 10 KO. Data are presented as mean ±SEM. *p<0.05. **p<0.01.
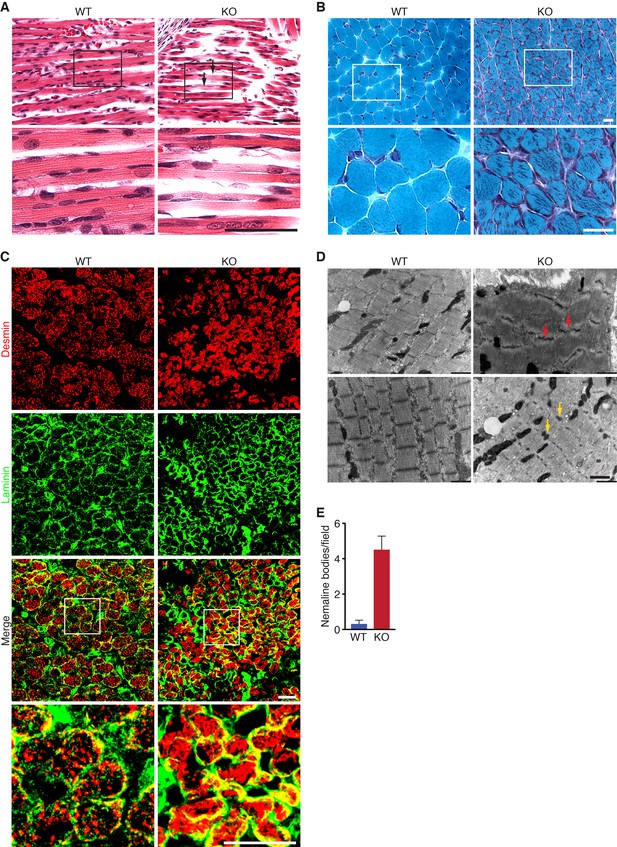
Loss of KLHL41 in mice leads to nemaline myopathy.
(A) Hematoxylin and eosin staining of longitudinal sections of diaphragm muscle of WT and Klhl41 KO mice at P0. Arrows point to ragged fibers (fibers with discontinuous H&E staining) present in KO muscle but not WT muscle. Bottom panels are zoomed images of the indicated regions. Scale bar: 20 µm. (B) Gomori’s trichrome staining of transverse sections of quadriceps muscle of WT and Klhl41 KO mice at P10. KO myofibers show numerous abnormal depositions absent in WT muscle. Bottom panels are zoomed images of the indicated regions. Scale bar: 20 µm. (C) Desmin and laminin immunostaining of transverse sections of WT and Klhl41 KO diaphragm at P3. Bottom panels are zoomed images of the indicated regions. Scale bar: 20 µm. (D) Electron microscopy images of WT and Klhl41 KO diaphragm at P3. Z-line streaming and nemaline bodies are indicated with red and yellow arrows, respectively. Scale bar: 1 µm. (E) Quantification of nemaline bodies from EM sections of WT and Klhl41 KO diaphragm at P3. The total number of electrondense protein aggregates (nemaline bodies) was counted for each field of view in WT and KO images (n = 8 fields per genotype). Data are presented as mean ±SEM. p<0.05.
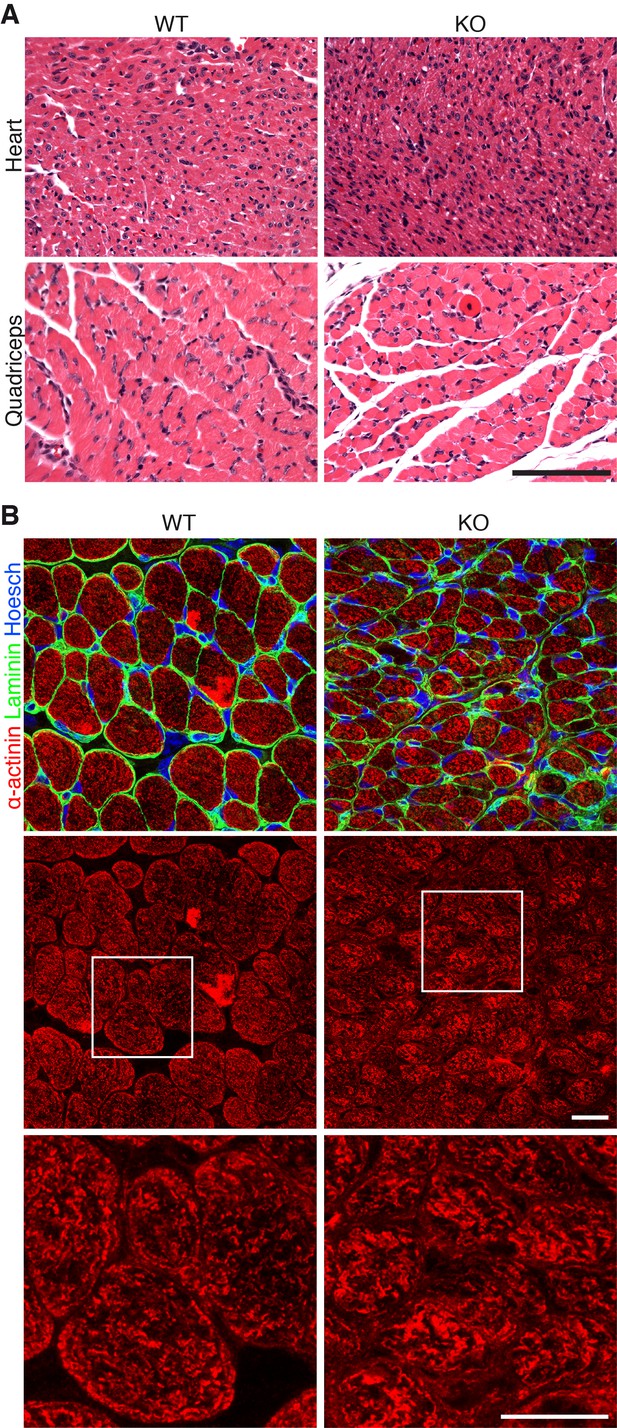
Additional histology of Klhl41 KO mice.
(A) Hematoxylin and eosin staining of transverse sections of heart ventricle (top) and quadriceps (bottom) of WT and Klhl41 KO mice at P3. Scale bar: 100 µm. (B) Sarcomeric α-actinin immunostaining of transverse sections of quadriceps of WT and Klhl41 KO mice at P10. Bottom panels are zoomed images of the indicated regions. Scale bar: 10 µm.
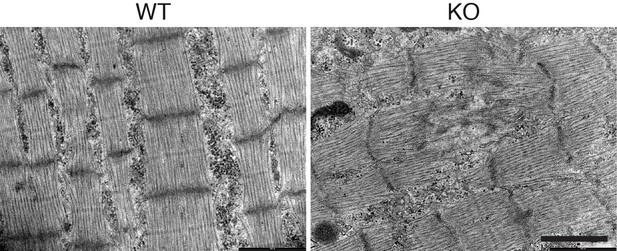
Ultrastructural abnormalities in Klhl41 KO hindlimb.
EM of WT and Klhl41 KO hindlimb muscle at P3. Scale bar: 1 µm.
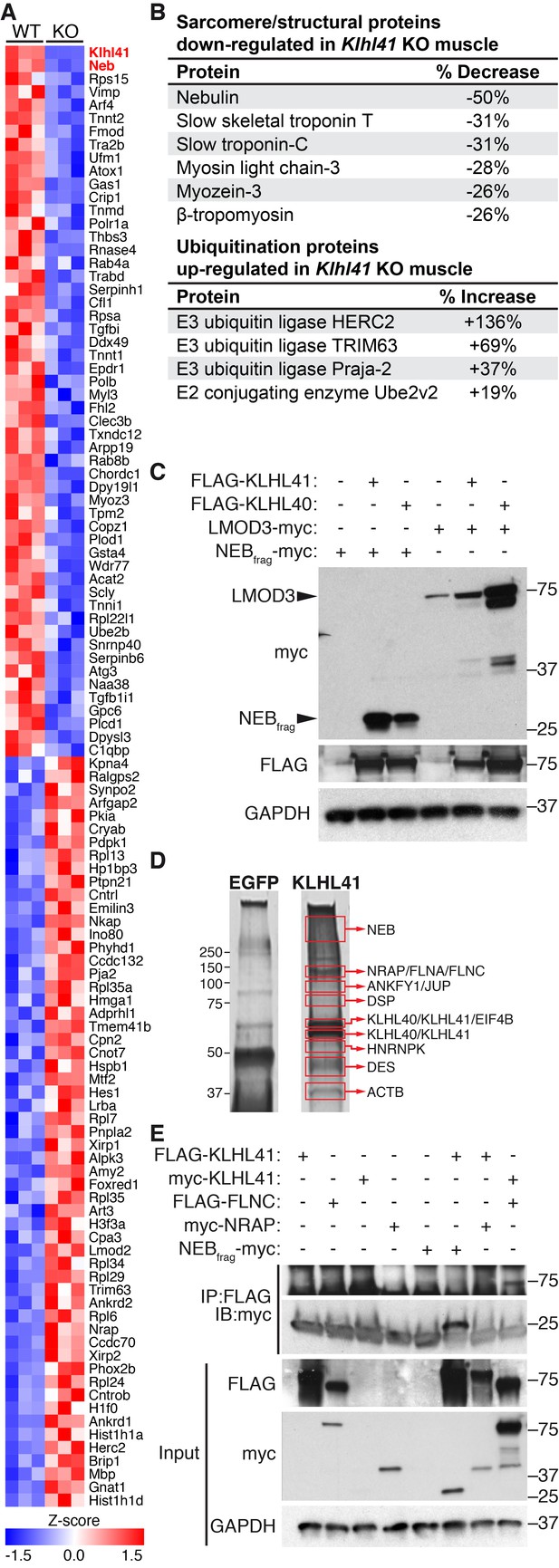
KLHL41 is required to maintain normal levels of sarcomeric proteins in vivo.
(A) Heat map of changes in protein levels identified by proteomics in WT and Klhl41 KO hindlimb muscles at P0 (n = 3 mice per genotype). A total of 389 proteins were identified by proteomics to be differentially regulated (p<0.05). Proteins up or down-regulated by more than 30% compared to WT are represented (n = 111). KLHL41 and NEB (red) were the two most down-regulated proteins. Heat map was created with Morpheus and the color scale represents Z-score. (B) Selected list of down-regulated sarcomeric proteins and up-regulated ubiquitination proteins identified by proteomics. Their relative levels to WT are shown on the right. (C) Effect of KLHL41 on NEBfrag and LMOD3 stability. KLHL41 preferentially stabilizes NEBfrag. COS-7 cells were transfected with expression vectors for the indicated proteins. Protein levels of LMOD3, NEBfrag, KLHL40 and KLHL41 were detected by western blot. GAPDH was used as a loading control. (D) KLHL41 binding partners identified following tandem affinity purification (TAP) from cultured C2C12 myotubes. Representative silver-stained gels with TAP protein from myotubes infected with 3XFLAG-HA-EGFP (negative control) or 3xFLAG-HA-KLHL41 are shown. Proteins listed next to each box indicate the most abundant protein(s) identified in each corresponding area. (E) Co-immunoprecipitation experiments to validate interactions between KLHL41 and FLNC, NRAP and NEBfrag. COS-7 cells were transfected with the indicated plasmids.
-
Figure 4—source data 1
Global protein changes in WT and KO mice by quantitative proteomics.
Unbiased quantitative proteomics was performed in WT and Klhl41 KO hindlimb muscle at P0 (n = 3 per genotype). Muscle samples were labeled with tandem mass tags (TMT). TMT129-131 were used to label WT samples and TMT126-128 were used to label KO samples. Data are provided as protein intensities (blue columns): for each channel, the signal of each unique peptide was added and normalized to the 126 channel. Then, protein intensities were generated by summing all of the unique peptide intensities to the corresponding protein. Proteins significantly regulated (p<0.05 and fold change >1.3) are shown. For up-regulated proteins, fold change was calculated as KOmean signal/WTmean signal. For down-regulated proteins, fold change was calculated as -WTmean signal/KOmean signal.
- https://doi.org/10.7554/eLife.26439.013
-
Figure 4—source data 2
Global mRNA changes in WT and KO mice by RNA-seq.
RNA-seq (n = 3 mice per genotype) in WT and KO hindlimb muscle at P0 was performed by the UT Southwestern Genomic and Microarray Core Facility. Data are provided as log of fold change (log(2)FC) and log2 of counts per million (log(2)CPM). For analysis of multiple groups, Holm-Sidak correction for multiple comparisons was utilized with a false discovery rate of 0.05 (FDR). Gene transcripts with significant changes (FDR < 0.05 and fold change >1.5) are included.
- https://doi.org/10.7554/eLife.26439.014
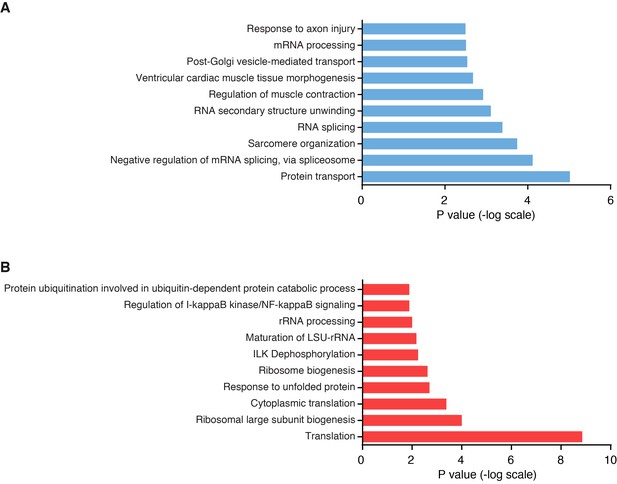
Pathway analysis of differentially regulated proteins in Klhl41 KO muscle by unbiased proteomics.
(A) Top 10 biological pathways enriched in down-regulated proteins in Klhl41 KO hindlimb muscle at P0 as identified by DAVID analysis. (B) Top 10 biological pathways enriched in up-regulated proteins in Klhl41 KO hindlimb muscle at P0 as identified by DAVID analysis.
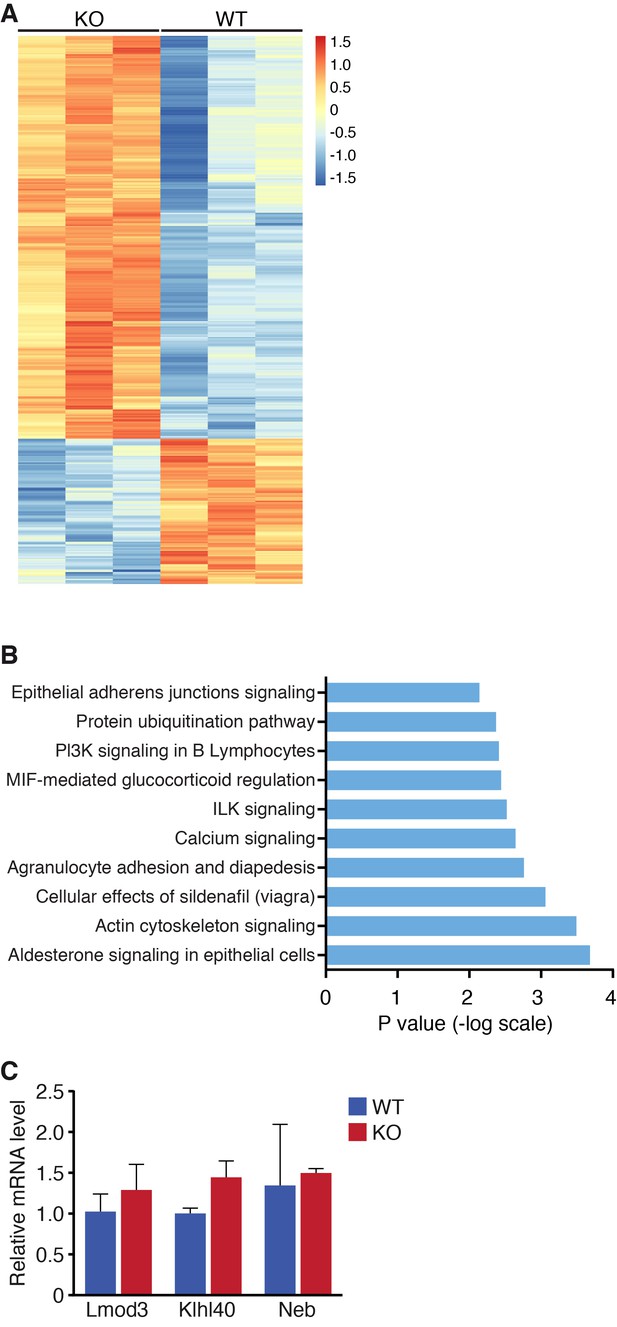
Global changes in mRNA expression by RNA-seq.
(A) Heat map of mRNA changes in WT and Klhl41 KO hindlimb muscle at P0 by RNA-seq (n = 3 per genotype). Data are represented as Z-score. (B) Ingenuity pathway analysis of up-regulated mRNA transcripts in Klhl41 KO hindlimb muscle at P0 identified by RNA-seq. (C) Detection of Lmod3, Klhl40 and Neb transcripts of WT and KO mice in P0 hindlimb muscle by qRT-PCR (n = 3 mice per genotype). Data are presented as mean ±SEM.
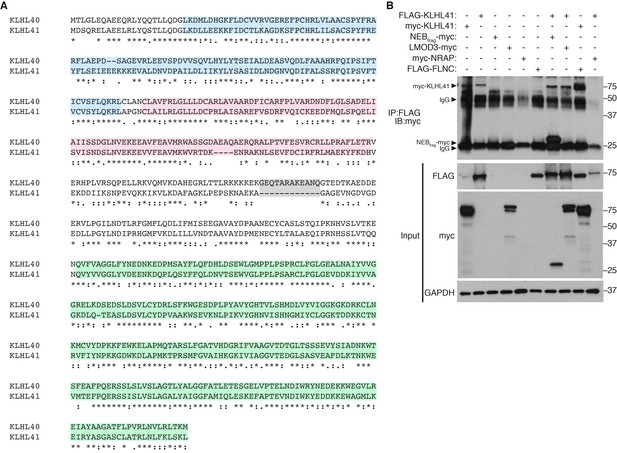
KLHL40 and KLHL41 show distinct partner specificity.
(A) Alignment between mouse KLHL40 and KLHL41 protein sequences. The different domains are indicated with distinct colors: BTB (blue), BACK (red) and Kelch repeats (green). Long insertions are highlighted in grey. (B) KLHL41 interacts with NEBfrag and FLNC but not LMOD3. Co-immunoprecipitation experiments were performed in COS-7 cells transfected with the indicated plasmids. Interaction between KLHL41 (75 kDa) and NEBfrag (27 kDa) and FLNC was detected in the pulldown (IP: FLAG IB:myc) but no specific band for LMOD3 (75 kDa) was observed. IgG bands from M2 anti-FLAG antibodies were detected at 25 kDa and 50 kDa.
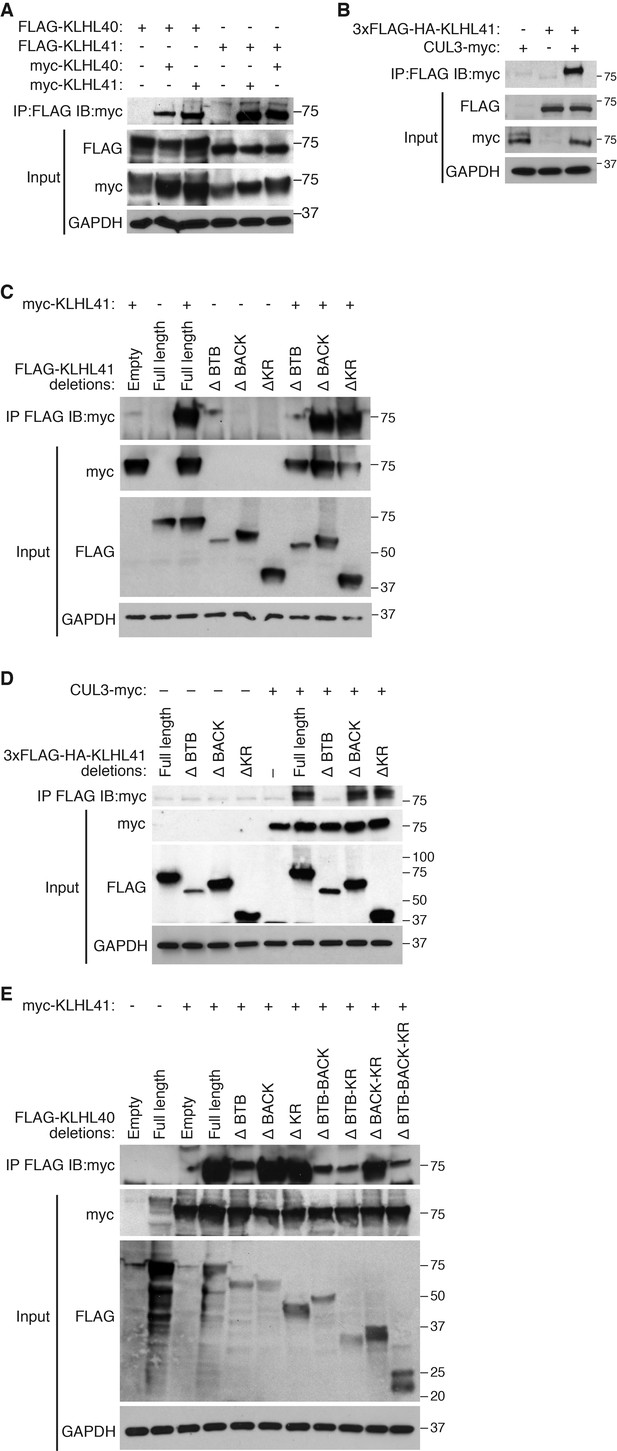
KLHL41 association with CUL3 is mediated by its BTB domain.
(A) Co-immunoprecipitation in transfected COS-7 cells indicates that KLHL41 homodimerizes with itself and heterodimerizes with KLHL40. Western blots of co-immunoprecipitation and input with indicated antibodies are shown. GAPDH was used as a loading control. (B) Interaction between 3XFLAG-HA-KLHL41 and CUL3-myc was detected by co-immunoprecipitation and western blot analysis with the indicated antibodies in transfected COS-7 cells. (C) Co-immunoprecipitation of various FLAG-tagged KLHL41 deletion mutants with full-length KLHL41 (myc-KLHL41) in transfected COS-7 cells to identify the domains necessary for KLHL41 self-dimerization. Deletion of the BTB domain (ΔBTB) abolished the interaction with full-length KLHL41. (D) Co-immunoprecipitation of various FLAG-tagged KLHL41 deletion mutants with CUL3-myc in transfected COS-7 cells to identify the domains necessary for association with CUL3. Deletion of the BTB domain (ΔBTB) abolished the interaction with CUL3-myc. (E) Co-immunoprecipitation of various FLAG-tagged KLHL40 deletion mutants with full-length KLHL41 (myc-KLHL41) in transfected COS-7 cells to identify the domains of KLHL40 required for heterodimerization with KLHL41. All FLAG-KLHL40 mutants in which the BTB domain of KLHL40 was deleted (ΔBTB; ΔBTB-BACK; ΔBTB−KR and ΔBTB−BACK−KR) presented diminished interaction with the full-length myc-KLHL41.
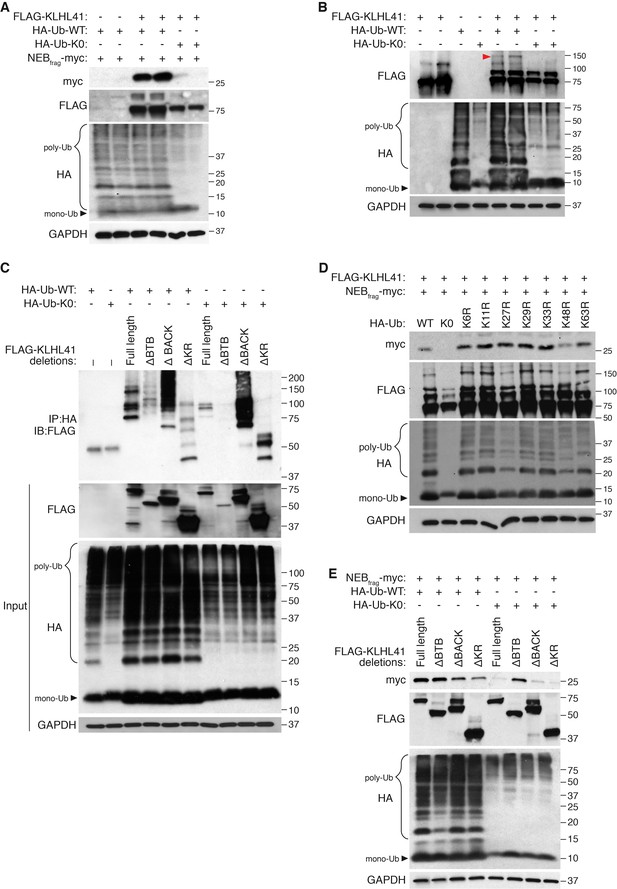
KLHL41 stabilization of NEBfrag is poly-ubiquitin-dependent.
(A) KLHL41 can stabilize NEBfrag in the presence of HA-Ub-WT but not HA-Ub-K0 mutant. Expression of NEBfrag and KLHL41 was determined by western blot analysis with the indicated antibodies in COS-7 cells. The laddered smear detected in HA input with HA-Ub-WT corresponds to the pool of poly-ubiquitinated proteins (indicated as poly-Ub). In contrast, HA-Ub-K0 (lysine zero), a mutant protein that cannot be poly-ubiquitinated, was preferentially detected as a single band of mono-Ubiquitin (indicated as mono-Ub). Note that HA-Ub-K0 prevented stabilization of NEBfrag by KLHL41. (B) Western blot analysis of FLAG-KLHL41 showed high molecular bands when expressed alone in COS-7 cells. Those bands were further increased when HA-Ub-WT was co-expressed (red arrowhead). Overexpression of HA-Ub-K0 collapsed the high molecular bands of FLAG-KLHL41, indicating that they correspond to a pool of poly-ubiquitinated KLHL41. (C) KLHL41 is preferentially poly-ubiquitinated in the BTB domain. Full-length and deletion mutants of KLHL41 were co-expressed in the presence of HA-Ub-WT or HA-Ub-K0 in COS-7 cells. Protein extraction was performed in the presence of deubiquitinase inhibitors and protein expression was detected by western blot analysis using the indicated antibodies. The laddered pattern in the IP:HA IB:FLAG western blot corresponds to poly-ubiquitinated KLHL41. Note that either deletion of BTB (ΔBTB) or HA-Ub-K0 overexpression collapsed poly-ubiquitinated bands. (D) HA-Ub-K48R impairs KLHL41 activity to stabilize NEBfrag. KLHL41 stabilization of NEBfrag in the presence of Ubiquitin HA-Ub-WT, K0 and Ub lysine mutants were determined by western blot analysis. HA-Ub-K48R overexpression led to a decrease in poly-ubiquitinated KLHL41, as observed in the FLAG input western blot. Each Ub lysine mutant only inhibits one type of ubiquitination. Therefore, total poly-ubiquitination levels remain the same as HA-Ub-WT as detected in HA input western blot. (E) Deletion of the KLHL41 BTB domain (ΔBTB) can restore NEBfrag stability in the presence of Ub-K0 mutant. Western blot analysis shows NEBfrag stability in the presence of full length and deletion mutants of KLHL41 when co-expressed with either HA-Ub-WT or HA-Ub-K0 mutant in COS-7 cells.

KLHL40 stabilization of LMOD3 is independent of poly-ubiquitination.
The ability of KLHL40 to stabilize LMOD3 in the presence of HA-Ub (WT) or HA-Ub (K0) was determined by western blot analysis with the indicated antibodies. The laddered smear detected in HA input with HA-Ub-WT corresponds to the pool of poly-ubiquitinated proteins (indicated as poly-Ub). In contrast, HA-Ub-K0 (lysine zero), a mutant protein that cannot be poly-ubiquitinated, was preferentially detected as a single band of mono-Ubiquitin (indicated as mono-Ub). LMOD3 protein levels did not change when HA-Ub-K0 was co-expressed compared to HA-Ub-WT.
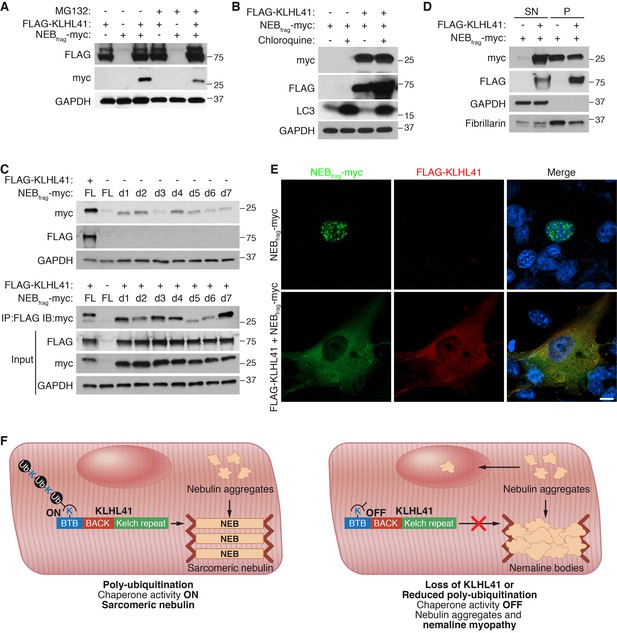
KLHL41 prevents NEBfrag aggregation in the nucleus.
(A) KLHL41 regulation of NEBfrag in the absence or presence of proteasome inhibitor (10 µM MG132 for 24 hr) was determined by western blot analysis with the indicated antibodies in COS-7 cells. Note that DMSO treated cells were used as negative (-) control. Expression of NEBfrag was not detected in the presence of MG132, and KLHL41 stabilizes NEBfrag in the presence or absence of MG132. (B) KLHL41 regulation of NEBfrag in the absence or presence of autophagy inhibitor (10 µM chloroquine for 12 hr) in COS-7 cells. Note that DMSO treated cells were used as negative (-) control. Expression of NEBfrag was not detected in the presence of chloroquine, and KLHL41 stabilizes NEBfrag in the presence or absence of chloroquine. Increased level of LC3 protein was detected as a positive control for autophagy inhibition. (C) Multiple NEBfrag deletion mutants are stable in the absence of KLHL41. COS-7 cells were transfected with various NEBfrag deletion mutants in the absence (top) or presence (bottom) of KLHL41. Protein levels were determined by western blot analysis with the indicated antibodies. Co-immunoprecipitation of FLAG-KLHL41 and NEBfrag deletion mutants (bottom) shows that all mutants interact with KLHL41. (D) Expression of KLHL41 affects NEBfrag solubility. COS-7 cells were transfected with the indicated plasmids and lysed with mild detergent to collect supernatant (SN). The remaining pellet (P) was then solubilized under high detergent conditions. Expression of FLAG-KLHL41 and NEBfrag in SN and P fractions was detected by western blot analysis. In the absence of KLHL41, NEBfrag was exclusively expressed in P. However, in the presence of KLHL41, total NEBfrag levels were increased and enriched in the SN fraction. GAPDH was restricted to the SN fraction. Fibrillarin, a nucleolar protein, was enriched in P. (E) Expression of NEBfrag in the absence and presence of KLHL41 was detected by immunofluorescence of COS-7 cells transfected with the indicated plasmids. Note that NEBfrag was accumulated in the nucleus in the absence of KLHL41 but it was redistributed to the cytoplasm when KLHL41 was co-expressed. Scale bar: 10 µm. (F) Model for regulation of KLHL41 stabilizing activity. KLHL41 acts as a poly-ubiquitin dependent chaperone that prevents NEB aggregation. Loss of KLHL41 or inhibition of its poly-ubiquitination leads to NEB aggregation, sarcomere disarray and nemaline myopathy.
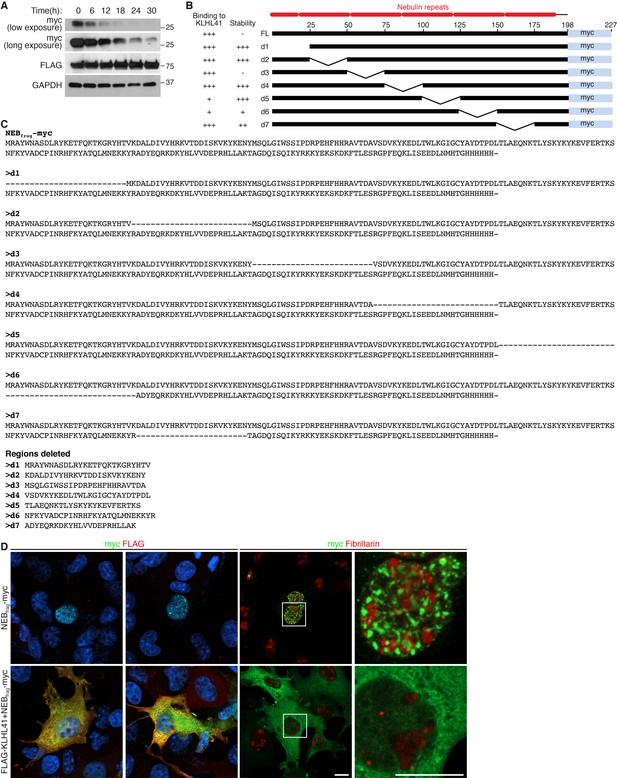
KLHL41 prevents NEBfrag aggregation in the nucleus.
(A) Determination of NEBfrag half-life. COS-7 cells transfected with NEBfrag-myc and FLAG-KLHL41 were pulsed with cycloheximide (100 µg/ml) for the indicated times and protein levels were determined by western blot analysis. GAPDH and KLHL41 protein levels did not change significantly with the treatment used. (B) Schematic of NEBfrag deletion mutants. NEBfrag is comprised of multiple nebulin repeats. Deletion mutants span 25 residues across the entire protein. The numbers on top of the full length (FL) sequence indicate the residue position (right). A summary of their binding to KLHL41 and stability in the absence of KLHL41 from Figure 7C is presented to the left. (C) Sequence alignment of NEBfrag deletion mutants (top). The sequence deleted in each mutant is also shown (bottom). (D) Immunofluorescence images showing NEBfrag localization in transfected COS-7 cells in the presence or absence of KLHL41. Fibrillarin was used as a nucleolar marker. The right panels are zoomed images of the indicated regions. Scale bar: 10 µm.
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.26439.020





