Science Forum: The Human Cell Atlas
Figures
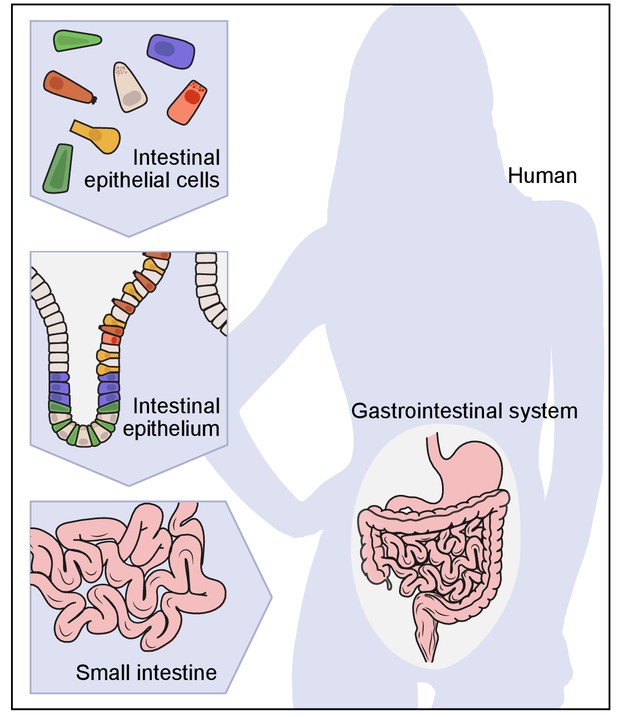
A hierarchical view of human anatomy.
A graphical depiction of the anatomical hierarchy from organs (such as the gut), to tissues (such as the epithelium in the crypt in the small intestine), to their constituent cells (such as epithelial, immune, stromal and neural cells).
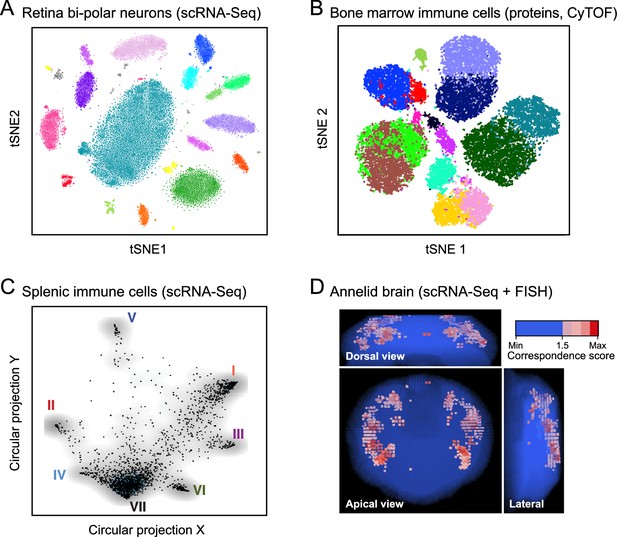
Anatomy: cell types and tissue structure.
The first three plots show single cells (dots) embedded in low-dimensional space based on similarities between their RNA-expression profiles (A, C) or protein-expression profiles (B), using either t-stochastic neighborhood embedding (A,B) or circular projection (C) for dimensionality reduction and embedding. (A) Bi-polar neurons from the mouse retina. (B) Human bone marrow immune cells. (C) Immune cells from the mouse spleen. (D) Histology. Projection of single-cell data onto tissue structures: image shows the mapping of individual cells onto locations in the marine annelid brain, based on the correspondence (color bar) between their single-cell expression profiles and independent FISH assays for a set of landmark transcripts.
© 2016 Elsevier Inc. Figure 2A reprinted from Shekhar et al., 2016 with permission.
© 2015 Elsevier Inc. Figure 2B reprinted from Levine et al., 2015 with permission.
© 2014 AAAS. Figure 2C reprinted from Jaitin et al., 2014 with permission.
© 2015 Macmillan Publishers Limited. Figure 2D adapted from Achim et al., 2015 with permission.
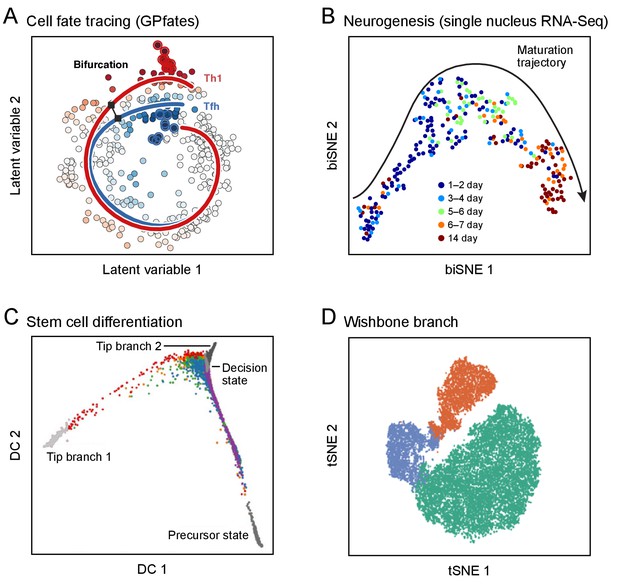
Developmental trajectories.
Each plot shows single cells (dots; colored by trajectory assignment, sampled time point, or developmental stage) embedded in low-dimensional space based on their RNA (A-C) or protein (D) profiles, using different methods for dimensionality reduction and embedding: Gaussian process patent variable model (A); t-stochastic neighborhood embedding (B, D); diffusion maps (C). Computational methods then identify trajectories of pseudo-temporal progression in each case. (A) Myoblast differentiation in vitro. (B) Neurogenesis in the mouse brain dentate gyrus. (C) Embryonic stem cell differentiation in vitro. (D) Early hematopoiesis.
© 2017 AAAS. Figure 3A reprinted from Lönnberg et al., 2017 with permission.
© 2016 AAAS. Figure 3B reprinted from Habib et al., 2016a with permission.
© 2016 Macmillan Publishers Limited. Figure 3C adapted from Haghverdi et al., 2016 with permission.
© 2016 Macmillan Publishers Limited. Figure 3D adapted from Setty et al., 2016 with permission.
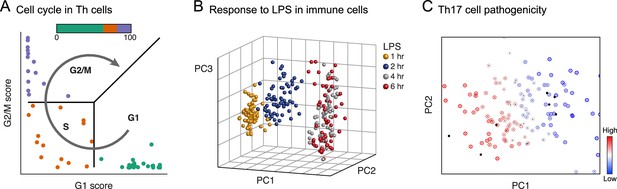
Physiology.
Each plot shows single cells (dots) embedded in low-dimensional space on the basis of their RNA profile, based on predefined gene signatures (A) or PCA (B, C), highlighting distinct dynamic processes. (A) The cell cycle in mouse hematopoietic stem and progenitor cells; adapted under terms of CC BY 4.0 from Scialdone et al. (2015). (B) Response to lipopolysaccharide (LPS) in mouse immune dendritic cells. (C) Variation in the extent of pathogenicity in mouse Th17 cells.
© 2014 Macmillan Publishers Limited. Figure 4B adapted from Shalek et al., 2014 with permission.
© 2015 Elsevier Inc. Figure 4C reprinted from Gaublomme et al., 2015 with permission.





