Immunoproteasome functions explained by divergence in cleavage specificity and regulation
Figures
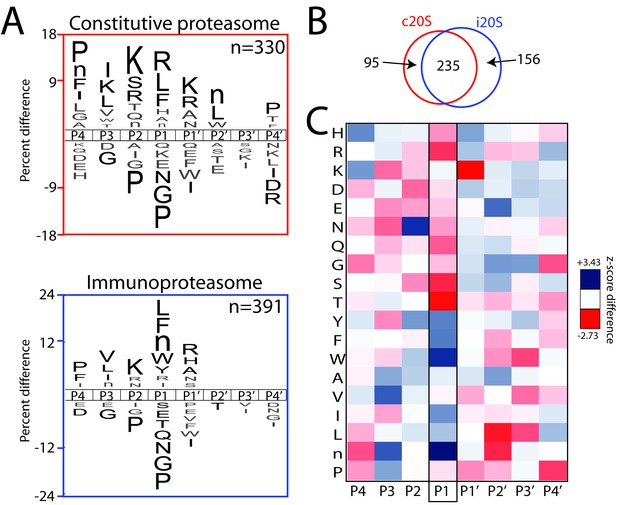
Global substrate specificity profiling of the iP and cP with Multiplex Substrate Profiling by Mass Spectrometry (MSP-MS) reveals shared and differential substrate specificity features.
(A) iceLogo representations of iP and cP substrate specificity (P4–P4ʹ) at the 480 min assay time point (p≤0.05 for non-grayed residues (Colaert et al., 2009); ‘n’ is norleucine). (B) Quantification of the total shared and non-overlapping iP- and cP-derived cleavages in the peptide library at the 480 min assay time point. Venn diagrams for additional assay time points are provided in Figure 1—figure supplement 1, demonstrating a time-dependent increase in cleavage overlap. (C) Heat map representation of iP and cP specificity differences using Z-scores (Colaert et al., 2009) calculated for the P4-P4ʹ positions. Differences in P1 specificity are highlighted. Heat maps for additional time points are provided in Figure 1—figure supplement 3. MSP-MS revealed that the iP has an increased preference for certain bulky, hydrophobic amino acid residues at the P1 position, whereas the cP has an increased preference for smaller and polar amino acid residues at the P1 position. A qualitative comparison of the P1 specificity differences identified using the MSP-MS library with those reported in Toes et al., 2001 and Mishto et al., 2014 is provided in Figure 1—figure supplement 4. Two biological replicates were assayed for each proteasome and only overlapping cleavages between replicates are reported. An analysis of biological (Figure 1—figure supplement 1) and technical (Supplementary file 1) reproducibility is provided.
-
Figure 1—source data 1
The following are contained in a supplementary file for the MSP-MS assay: a sample key; the sequences of the peptide library in FASTA format; a full mass spectrometry peptide report; processed cleavage data showing spectral counts for each octapeptide (P4–P4ʹ) and the associated cleavage within the parental peptide; processed cleavage data (indicated as ‘negative’) with cleavages also appearing in the no-enzyme control (NEC) highlighted; all octapeptides (P4–P4ʹ) to be used as the background dataset in iceLogo; ‘positive’ octapeptide (P4–P4ʹ) cleavages with the ‘negative’ cleavage data removed; ‘accumulative’ octapeptide (P4–P4ʹ) cleavages at each assay time point for the proteasome biological replicates; a comparison of cleavages common between biological replicates; and heat maps based on Z-scores showing the specificity at each assay time point.
- https://doi.org/10.7554/eLife.27364.007
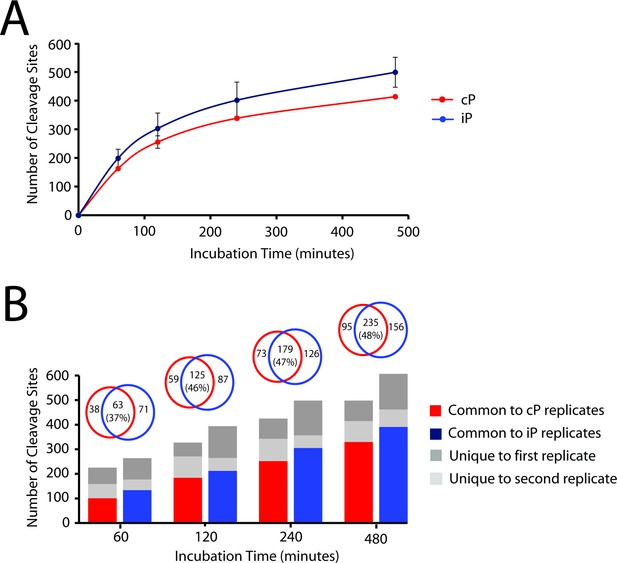
Comparison of iP- and cP-derived peptide cleavage products across all time points in the MSP-MS assay.
(A) Progress curves for the iP and cP are shown demonstrating the kinetics of peptide cleavage during the MSP-MS assay time course. (B) Quantification of shared and non-overlapping cleavages is provided between two biological replicates of each proteasome (bar graph). Further comparison of iP and cP cleavage overlap is shown using only cleavages observed in both biological replicates (Venn diagrams). This analysis revealed that the iP and cP display a time-dependent increase in the number of shared cleavages.
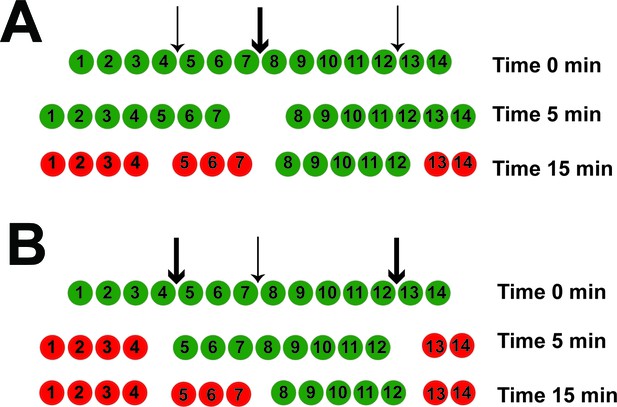
The MSP-MS assay detects the most dominant peptide cleavage events.
These cleavage events are those that tend to occur initially and produce peptides of appropriate size (in m/z). For a given protease, the assay is capable of detecting up to approximately 20% of the total peptide bonds that can be cleaved in the library, which is similar to the maximum cleavage-site usage observed for each proteasome (Figure 1—figure supplement 1). In this illustrated example, green peptides are detected, whereas red peptides cannot be detected. The cP (A) and iP (B) cleave one of the 14-mer peptides at three positions, between residues 4–5, 7–8, and 12–13. If cleavage at the 7–8 position (thick arrow) by the cP is much faster than the 4–5 and 12–13 cleavages (thin arrows), then the 4–5 cleavage product is not detected. However, for the iP, if the 4–5 and 12–13 cleavages are much faster than the 7–8 cleavage, then all three sites can be detected, and the 4–5 cleavage site would be reported as unique to the iP.
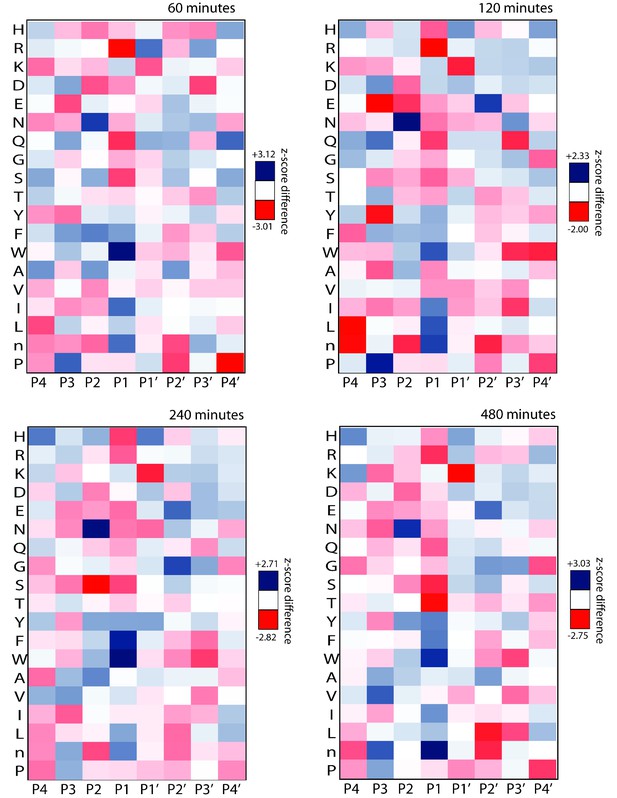
Heat maps for the iP and cP across multiple MSP-MS assay time points demonstrating shared and differential specificity features at the P4 to P4ʹ positions.
The heat map from the 480 min assay time point has been repeated from Figure 1 for clarity.
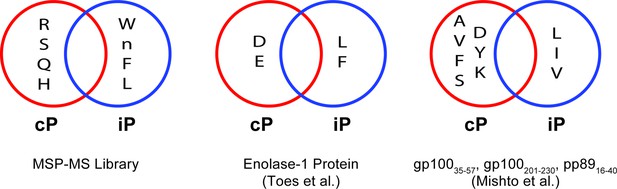
Qualitative comparison of the major differences in human iP and cP P1 specificity preferences identified using the MSP-MS library compared to those reported in previous profiling studies.
Toes et al used a model protein substrate (enolase-1) for proteasome cleavage, (Toes et al., 2001) whereas Mishto et al used a set of synthetic peptides corresponding to the sequences gp10035-57, gp100201-230, and pp8916-40 (Mishto et al., 2014).
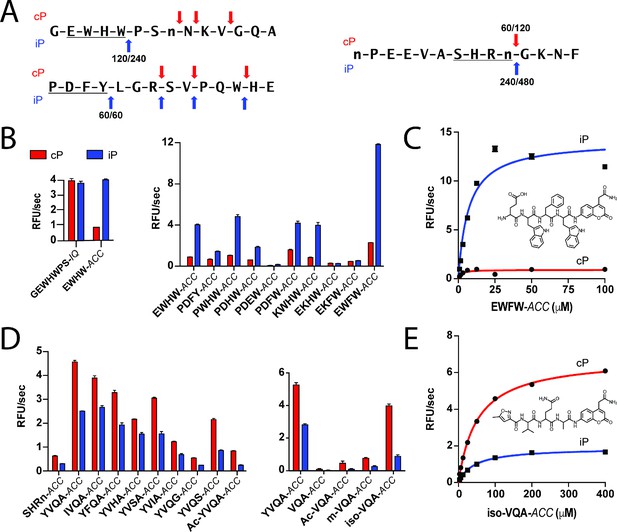
Rational optimization of tetrapeptide ACC substrates with iP and cP selectivity.
(A) Example 14-mer peptides from the iP and cP MSP-MS assays associated with Figure 1 illustrating cleavages that were used as a template for rational fluorogenic substrate design. All cleavages common between two biological replicates of each proteasome are reported with the time point of first appearance noted for each replicate. (B) Selectivity of parental IQ and tetrapeptide ACC fluorogenic substrates derived from an iP-favored cleavage (Figure 2A, top) in the MSP-MS library. Subsequent rational substrate optimization is shown. (C) Michaelis-Menten characterization and chemical structure of the lead iP substrate, EWFW-ACC. (D) Optimization of an ACC substrate with cP selectivity using a similar approach. Shortening the peptide sequence to a tripeptide and addition of an N-terminal capping group were found to be important for improving substrate selectivity and maintaining specific activity. (E) Michaelis-Menten characterization and chemical structure of the lead cP substrate iso-VQA-ACC. For all fluorogenic substrate assays, mean activity is reported with error bars representing the standard deviation from n = 3 replicates.
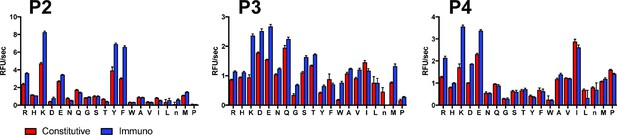
P4-P2 substrate specificity of SDS-activated iP and cP using the PS-SCL profiling approach.
Mean activity is reported with error bars representing the standard deviation from n = 3 replicates.
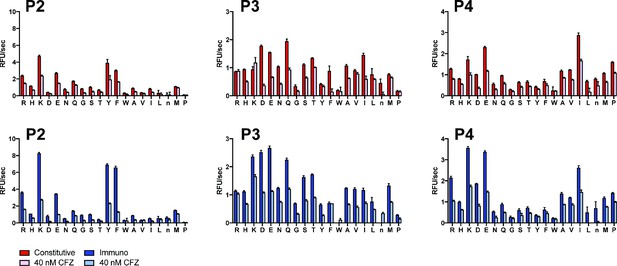
PS-SCL profiling of SDS-activated iP and cP in the absence and presence of 40 nM CFZ pretreatment.
CFZ levels were selected based on inhibitor titration against the library to achieve optimal selectivity. Mean activity is reported with error bars representing the standard deviation from n = 3 replicates.
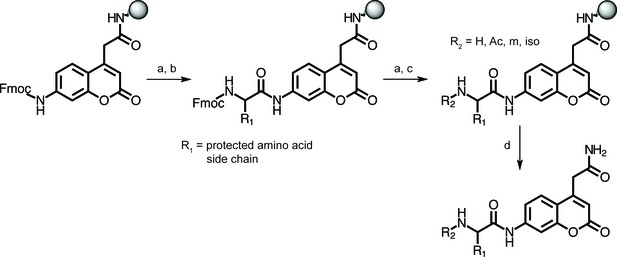
Synthetic approach for peptide substrates bearing a C-terminal ACC fluorophore.
Protocols for synthesis of the Fmoc-ACC fluorophore and its coupling to Rink amide resin have been described. (Maly et al., 2002) (a) 20% 4-methylpiperidine/DMF; (b) Fmoc-protected P1 amino acid (5 eq.), HATU (5 eq.), and 2,4,6-collidine (5 eq.), DMF, 24 hr, or Fmoc-protected P2, P3, or P4 amino acid (6.5 eq.), HBTU (6.5 eq.), and N-methylmorpholine (13 eq.), DMF, 1 hr; (c) for acetyl (Ac): acetic anhydride/TEA/DMF/DCM (1:1:5:5), for N-morpholinyl (m): 4-morpholinylacetic acid hydrochloride (6.5 eq.), HBTU (6.5 eq.), and N-methylmorpholine (26 eq.), and for 5-methylisoxazolyl (iso): 5-methylisoxazole-3-carboxylic acid (6.5 eq.), HBTU (6.5 eq.), and N-methylmorpholine (13 eq.); (d) TFA/H2O/triisopropylsilane (95:2.5:2.5).
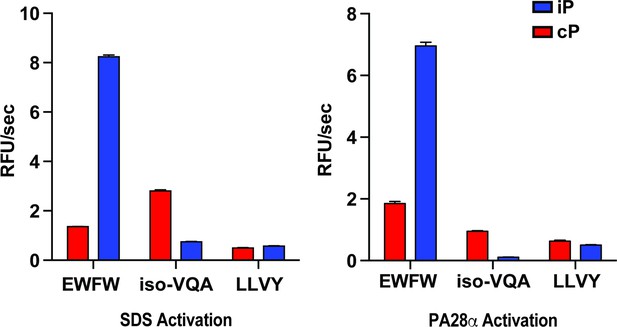
Comparison of fluorogenic substrate selectivity under SDS (0.03%) and PA28 (12 eq.) activation conditions.
Mean activity is reported with error bars representing the standard deviation from n = 3 replicates.
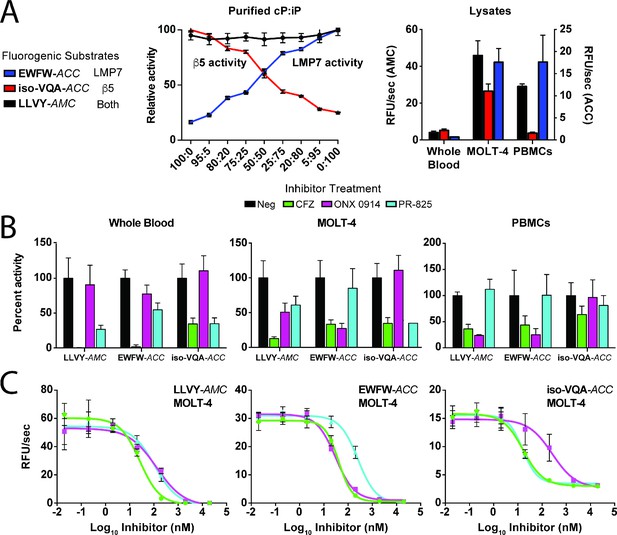
Optimal fluorogenic substrates enable the selective monitoring of LMP7 and β5 activity in cell lysates containing varied proteasome ratios.
(A) Demonstration of EWFW-ACC (10 μM) and iso-VQA-ACC (30 μM) selectivity for the iP and cP, respectively, compared to the universal substrate LLVY-AMC (30 μM) using mixed ratios of purified proteasomes. A comparison is provided of EWFW-ACC and iso-VQA-ACC specificity in whole blood, MOLT-4, and PBMC lysates. (B) Pretreatment of the lysates with 100 nM CFZ, an LMP7-favored inhibitor (ONX 0914), or a β5-favored inhibitor (PR-825) demonstrates the ability of the substrates to distinguish their target proteasomes under lysate conditions. (C) IC50 determination for CFZ, ONX 0914, and PR-825 in the mixed LMP7/β5 MOLT-4 lysate using LLVY-AMC and the selective substrates. For all assays, mean activity is reported with error bars representing the standard deviation from n = 3 replicates.
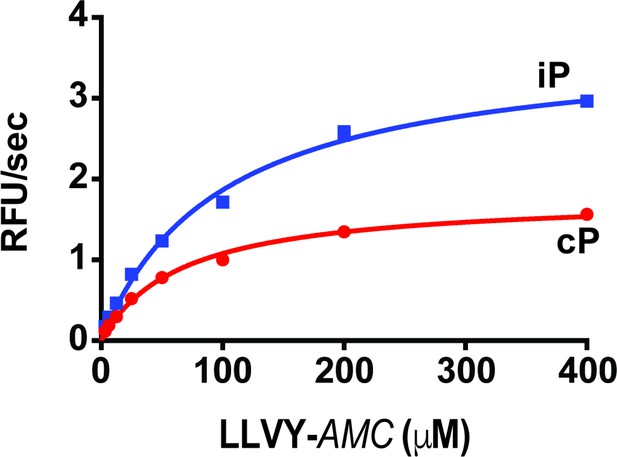
Michaelis-Menten plot of the iP and cP with the commercial LLVY-AMC substrate.
Km = 98.6 μM for the iP and 65.7 μM for the cP with 2.0 fold selectivity at Vmax.
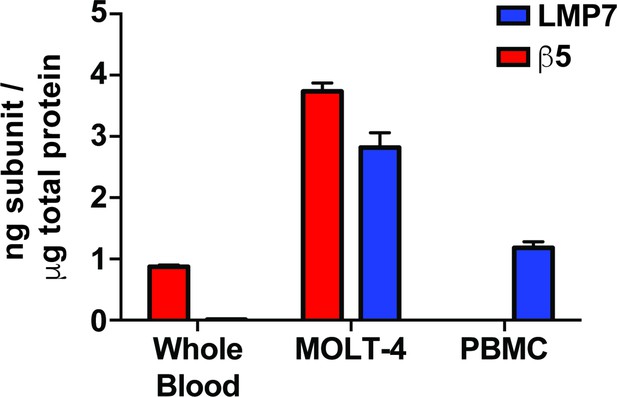
Quantification of active LMP7 and β5 levels in lysates from whole blood, the MOLT-4 cell line, and PBMCs through direct active site labeling using the ProCISE assay.
Mean labeled subunit levels are reported with error bars representing the standard deviation from n = 3 replicates.
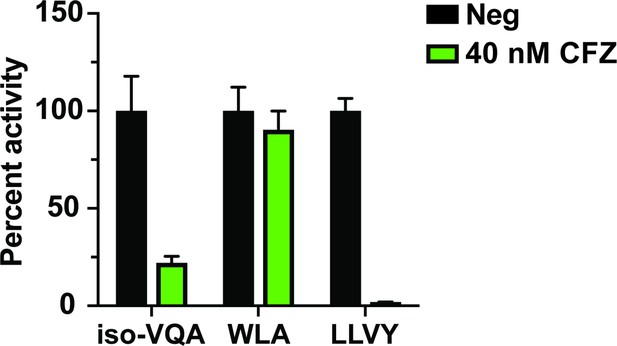
Improved ability of the iso-VQA-ACC substrate to detect β5 activity in the MOLT-4 lysate compared to a commercially available β5 fluorogenic substrate.
Mean activity is reported with error bars representing the standard deviation from n = 3 replicates.
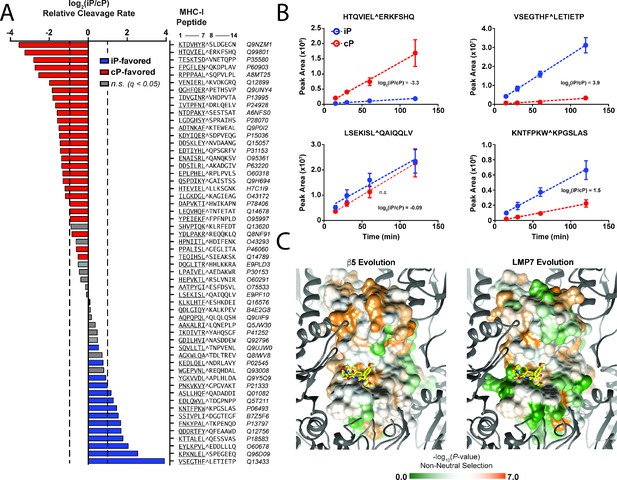
Differential substrate specificity of the iP alters MHC I peptide cleavage and is likely due to neutral evolution from the ancestral cP.
(A) Label-free quantitation of relative iP and cP cleavage rates against a library of synthetic peptides derived from sequences flanking sites of MHC I peptide processing (Bassani-Sternberg et al., 2015). Residues corresponding to the C-terminal portion of a given MHC I peptide are at positions 1–7 in the peptide library, whereas residues corresponding to the subsequent parent protein sequence are at positions 8–14. Average relative cleavage rates (log2) are provided for iP and cP substrates between positions 7 and 8 (P1/P1ʹ) for all peptides in the library that underwent cleavage at this site and for which quantification in all replicates was possible (n = 4). Two-fold differences in relative cleavage rate (log2 = 1) are indicated with a dashed line. Non-grayed bars represent statistically significant (q < 0.05) differences in selectivity as determined using a Student’s t-test (see Statistical methods). (B) Example kinetic traces from the MHC I peptide library time course showing cleavages following hydrophobic residues that have either high selectivity or no significant selectivity for the iP. Mean peak areas are reported with error bars representing the standard deviation (n = 4). (C) Evolutionary selection of residues in the β5 and LMP7 subunits across species that contain both proteasome isoforms. The β5 subunit has undergone more significant non-neutral evolution (residues are colored in orange) compared to the LMP7 subunit. Differences in evolutionary selection may account for the divergence in LMP7 and β5 cleavage specificity. Sequence alignments are provided in Figure 4—figure supplement 2.
-
Figure 4—source data 1
The following are contained in a supplementary file for the MHC cleavage assay: a sample key; the sequences of the peptide library in FASTA format; and a full mass spectrometry peptide report.
- https://doi.org/10.7554/eLife.27364.018
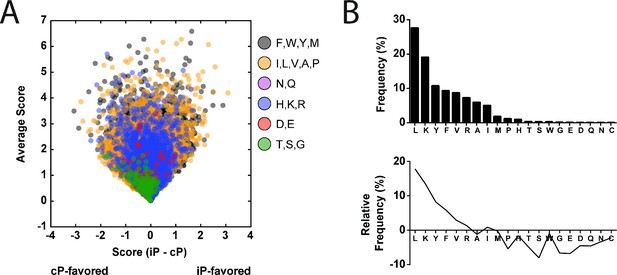
Prediction of MHC I peptide cleavage by the iP and cP.
(A) Scoring of the MHC I peptidome (Bassani-Sternberg et al., 2015) (n = 22,598) based on the relative favorability for cleavage by the iP or cP. Scores were derived from the P4-P1 cleavage preferences of each proteasome as determined in the MSP-MS assay. MHC I peptides are colored according to their C-terminal residue (cysteine was omitted because it is absent in the MSP-MS library; norleucine was incorporated into the MSP-MS library instead of methionine and was used for scoring methionine residues). (B, top) Frequency of the C-terminal residues in the MHC I peptidome. (B, bottom) Relative frequency of the C-terminal residues following subtraction of the frequencies of the corresponding amino acids in the human proteome (Liu et al., 2010b).
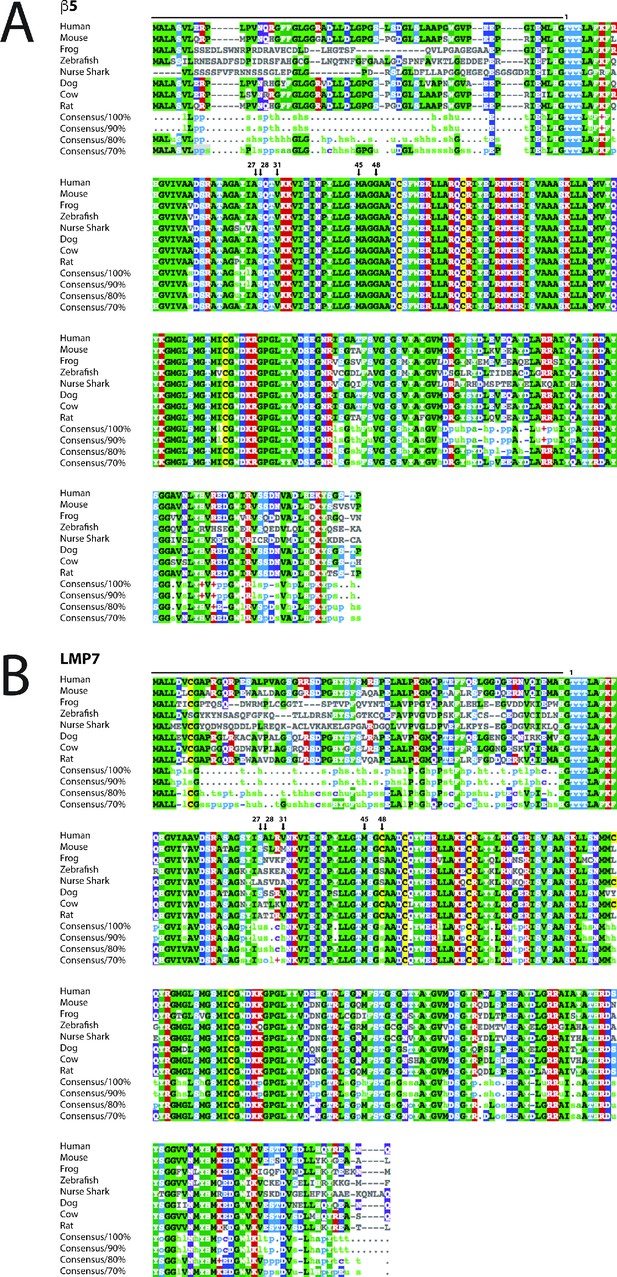
Protein sequence alignment for (A) β5 and (B) LMP7 in diverse species containing both proteasomes.
(Sutoh et al., 2012) UniProt identification numbers are respectively as follows for β5 and LMP7: P28074 and P28062, Homo sapiens (human); O55234 and P28063, Mus musculus (mouse); P28075 and P28064, Rattus norvegicus (rat); Q32KL2 and Q3T112, Bos taurus (bovine); E2R3R2 and Q5W416, Canis familiaris (dog); F6WGD3 and A8E5T8, Xenopus tropicalis (western clawed frog); A8E5G8 and O57330, Danio rerio (zebrafish); Q90572 and Q90571, Ginglymostoma cirratum (nurse shark). Key residues described in the text are labeled for comparison.
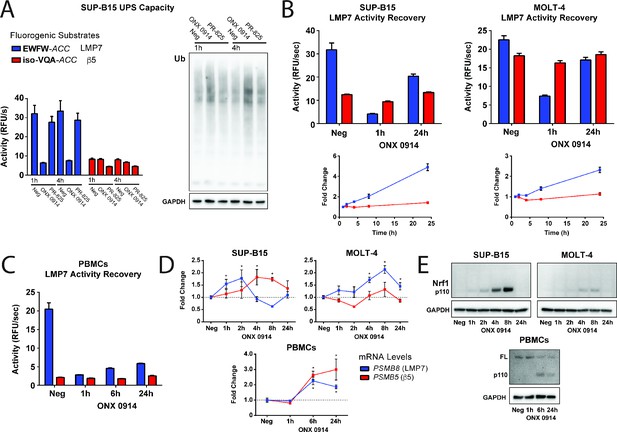
iP activity makes a significant contribution to UPS capacity and undergoes selective and coordinated recovery from inhibition.
(A) Ubiquitin western blot and matched activity assay for the SUP-B15 cell line (with approximately 80:20 LMP7: β5 activity) showing a substantial increase in global ubiquitination levels upon one-hour pulse treatment with ONX 0914. A significant increase in ubiquitination was not observed with PR-825 treatment, suggesting that LMP7 is a more dominant contributor to UPS capacity in this cell line in accordance with the proportionally higher LMP7 activity level. Selective recovery of LMP7 activity over 24 hr in the (B) SUP-B15 and MOLT-4 cell lines and in (C) primary PBMCs from a healthy donor (see also Figure 5—figure supplement 2) upon one-hour pulse treatment with ONX 0914. The SUP-B15 and MOLT-4 lower panels show the fold change in LMP7 and β5 activity levels normalized to the one-hour time point. For all activity assays, mean activity is reported with error bars representing the standard deviation from n = 3–6 replicates. (D) Corresponding qRT-PCR analysis of PSMB8 (LMP7) and PSMB5 (β5) mRNA dynamics in the cell lines and PBMCs following ONX 0914 treatment. Mean expression levels are reported with normalization to GAPDH. Statistically significant differences are indicated (*p≤0.05) as determined using a Student’s t-test with error bars representing the SEM from n = 3 replicates. (E) Corresponding western blots demonstrating accumulation of processed Nrf1 (p110) in RIPA lysates from the cell lines and PBMCs following ONX 0914 treatment. Full-length (FL) Nrf1 was observed in the PBMC lysates. .
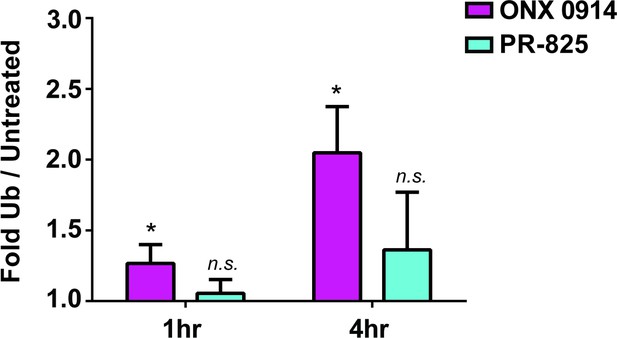
Quantification of the ubiquitin levels in the SUP-B15 cell line at the one- and four-hour time points following one-hour pulse treatment with ONX 0914 and PR-825 (Figure 5A).
Mean ubiquitin levels are reported with normalization to the untreated condition. Statistically significant differences compared to the untreated condition are indicated (*p≤0.05) as determined with a Student’s t-test with error bars representing the standard deviation from n = 3 replicates.
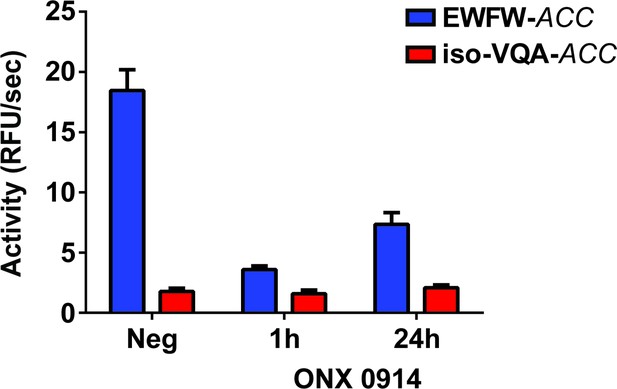
Recovery of LMP7 activity in PBMCs from n = 3 donors over 24 hr following one-hour pulse treatment with ONX 0914.
Mean activity is reported with error bars representing the standard deviation from n = 3 replicates per donor.
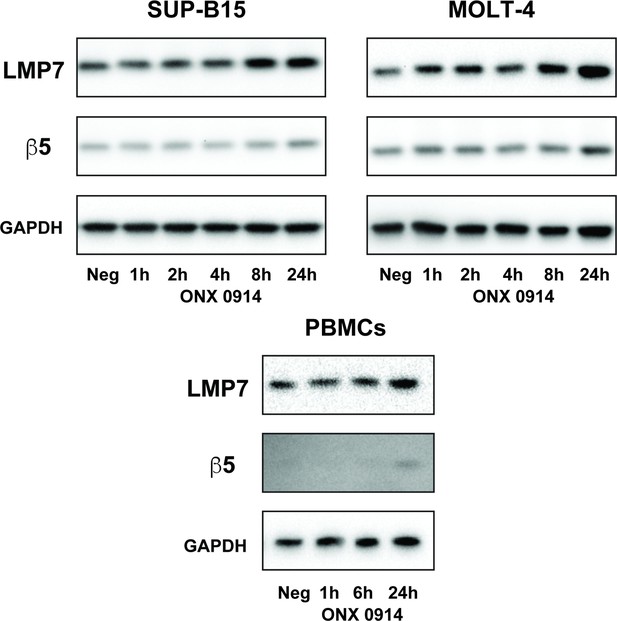
Western blots of LMP7 and β5 subunit levels during the ONX 0914 recovery time course in the SUP-B15 and MOLT-4 cell lines and in PBMCs from a healthy donor.
https://doi.org/10.7554/eLife.27364.024Additional files
-
Supplementary File 1
Comparison of the reproducibility of the MSP-MS assay for technical triplicate samples.
Percent identity was compared across replicates for the intact library (‘no enzyme control’) and after the addition of the immunoproteasome as an example. The 8 hr time point was chosen to maximize the number of cleavages in the comparison.
- https://doi.org/10.7554/eLife.27364.025
-
Supplementary File 2
Comparison of select ACC substrate Michaelis-Menten parameters.
- https://doi.org/10.7554/eLife.27364.026
-
Supplementary File 3
Comparison of compound IC50 values in the MOLT-4 lysate obtained using the optimal fluorogenic substrates and the ProCISE assay.
- https://doi.org/10.7554/eLife.27364.027
-
Transparent reporting form
- https://doi.org/10.7554/eLife.27364.028





